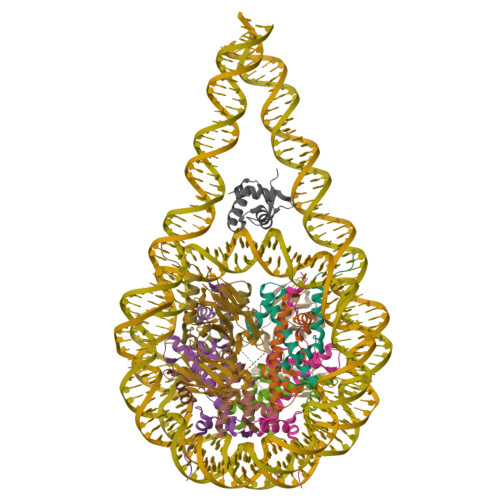
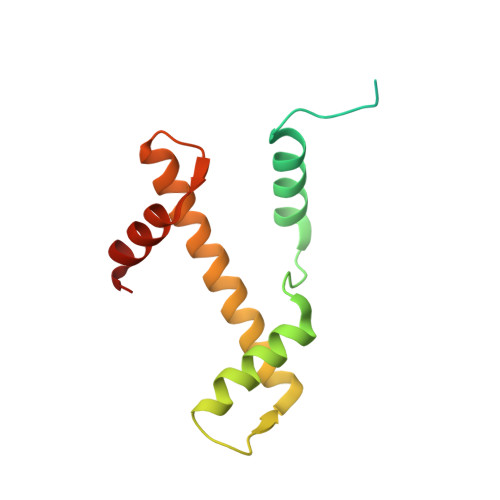
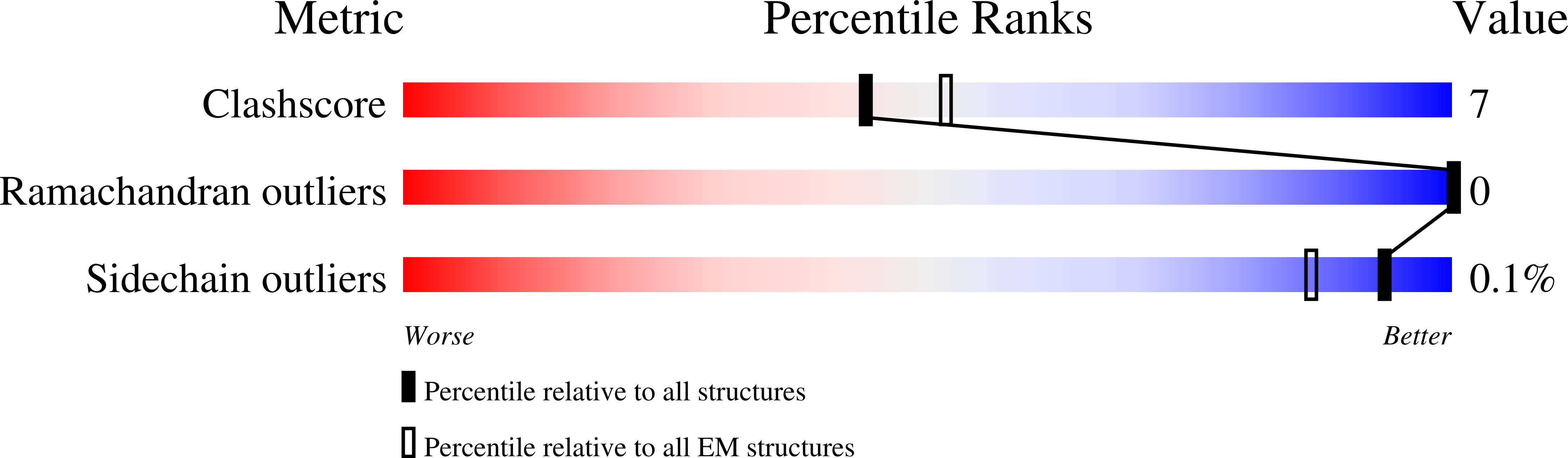Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms.
Zhou, B.R., Feng, H., Kale, S., Fox, T., Khant, H., de Val, N., Ghirlando, R., Panchenko, A.R., Bai, Y.(2021) Mol Cell 81: 166-182.e6
- PubMed: 33238161
- DOI: https://doi.org/10.1016/j.molcel.2020.10.038
- Primary Citation of Related Structures:
7K5X, 7K5Y, 7K60, 7K61, 7K63 - PubMed Abstract:
The repeating structural unit of metazoan chromatin is the chromatosome, a nucleosome bound to a linker histone, H1. There are 11 human H1 isoforms with diverse cellular functions, but how they interact with the nucleosome remains elusive. Here, we determined the cryoelectron microscopy (cryo-EM) structures of chromatosomes containing 197 bp DNA and three different human H1 isoforms, respectively. The globular domains of all three H1 isoforms bound to the nucleosome dyad. However, the flanking/linker DNAs displayed substantial distinct dynamic conformations. Nuclear magnetic resonance (NMR) and H1 tail-swapping cryo-EM experiments revealed that the C-terminal tails of the H1 isoforms mainly controlled the flanking DNA orientations. We also observed partial ordering of the core histone H2A C-terminal and H3 N-terminal tails in the chromatosomes. Our results provide insights into the structures and dynamics of the chromatosomes and have implications for the structure and function of chromatin.
Organizational Affiliation:
Laboratory of Biochemistry and Molecular Biology, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.