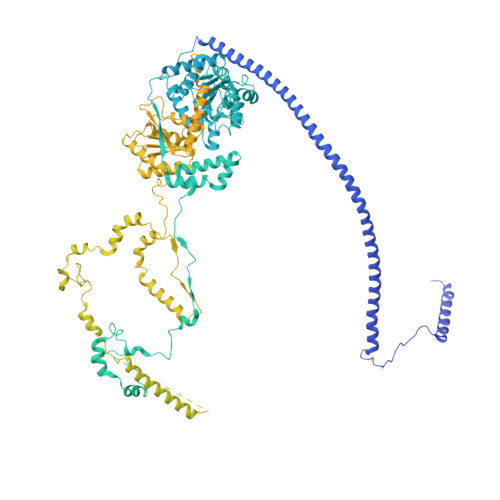
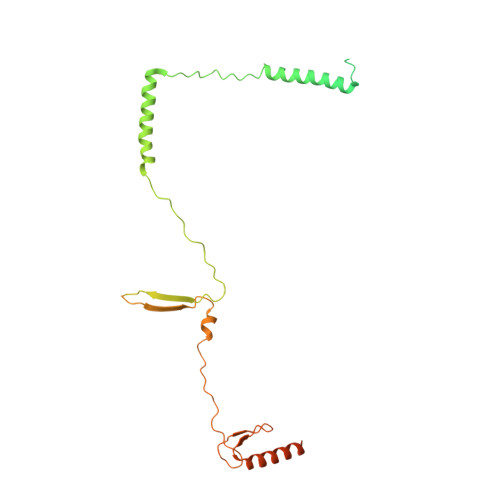
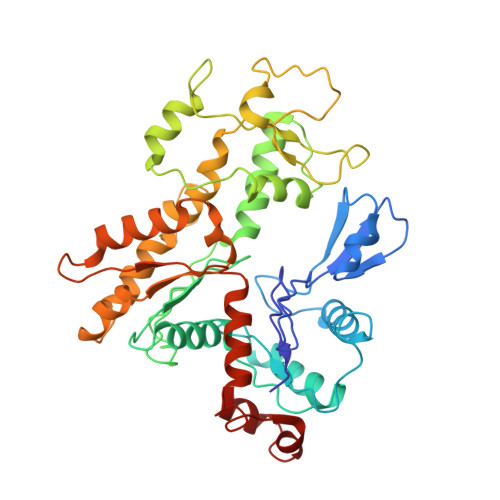
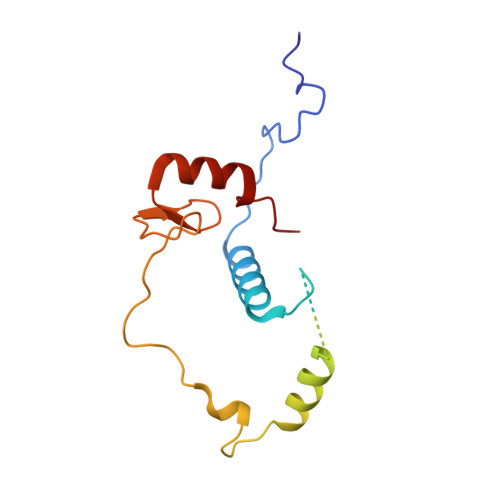
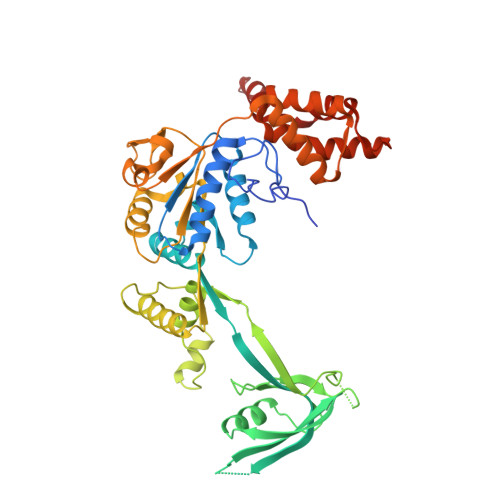
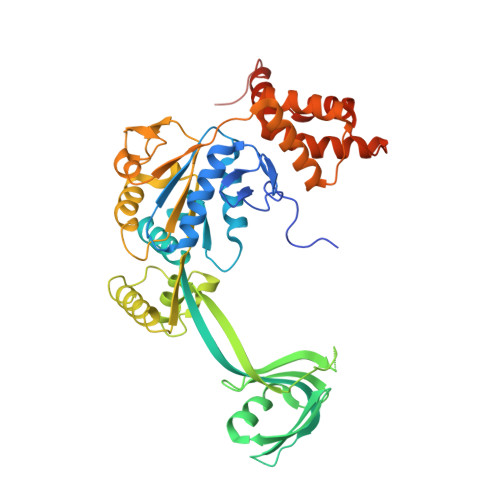
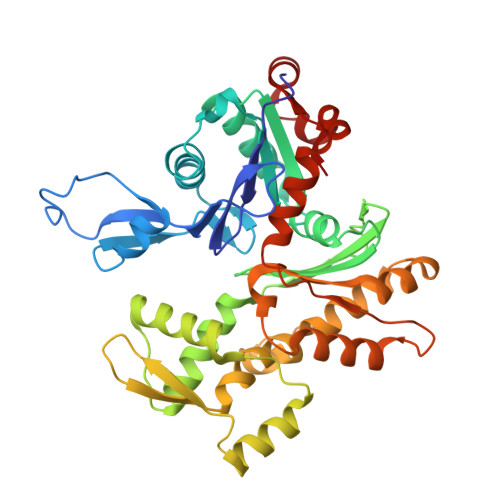
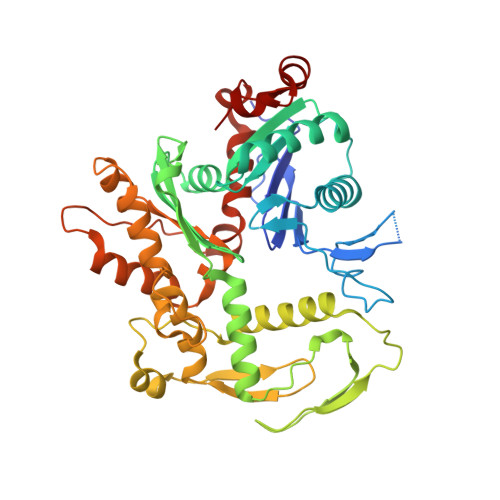
Structural insights into histone exchange by human SRCAP complex.
Yu, J., Sui, F., Gu, F., Li, W., Yu, Z., Wang, Q., He, S., Wang, L., Xu, Y.(2024) Cell Discov 10: 15-15
- PubMed: 38331872
- DOI: https://doi.org/10.1038/s41421-023-00640-1
- Primary Citation of Related Structures:
8X15, 8X19, 8X1C - PubMed Abstract:
Histone variant H2A.Z is found at promoters and regulates transcription. The ATP-dependent chromatin remodeler SRCAP complex (SRCAP-C) promotes the replacement of canonical histone H2A-H2B dimer with H2A.Z-H2B dimer. Here, we determined structures of human SRCAP-C bound to H2A-containing nucleosome at near-atomic resolution. The SRCAP subunit integrates a 6-subunit actin-related protein (ARP) module and an ATPase-containing motor module. The ATPase-associated ARP module encircles half of the nucleosome along the DNA and may restrain net DNA translocation, a unique feature of SRCAP-C. The motor module adopts distinct nucleosome binding modes in the apo (nucleotide-free), ADP-bound, and ADP-BeF x -bound states, suggesting that ATPase-driven movement destabilizes H2A-H2B by unwrapping the entry DNA and pulls H2A-H2B out of nucleosome through the ZNHIT1 subunit. Structure-guided chromatin immunoprecipitation sequencing analysis confirmed the requirement of H2A-contacting ZNHIT1 in maintaining H2A.Z occupancy on the genome. Our study provides structural insights into the mechanism of H2A-H2A.Z exchange mediated by SRCAP-C.
- Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, New Cornerstone Science Laboratory, State Key Laboratory of Genetic Engineering and Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College of Fudan University, Shanghai, China.
Organizational Affiliation: