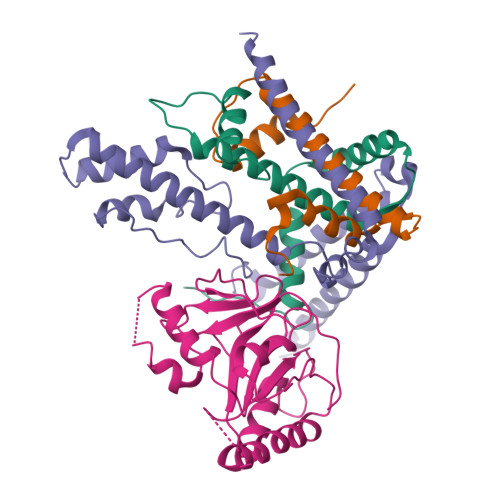
Structural basis underlying viral hijacking of a histone chaperone complex.
Huang, H., Deng, Z., Vladimirova, O., Wiedmer, A., Lu, F., Lieberman, P.M., Patel, D.J.(2016) Nat Commun 7: 12707-12707
- PubMed: 27581705
- DOI: https://doi.org/10.1038/ncomms12707
- Primary Citation of Related Structures:
5KDM - PubMed Abstract:
The histone H3.3 chaperone DAXX is implicated in formation of heterochromatin and transcription silencing, especially for newly infecting DNA virus genomes entering the nucleus. Epstein-Barr virus (EBV) can efficiently establish stable latent infection as a chromatinized episome in the nucleus of infected cells. The EBV tegument BNRF1 is a DAXX-interacting protein required for the establishment of selective viral gene expression during latency. Here we report the structure of BNRF1 DAXX-interaction domain (DID) in complex with DAXX histone-binding domain (HBD) and histones H3.3-H4. BNRF1 DID contacts DAXX HBD and histones through non-conserved loops. The BNRF1-DAXX interface is responsible for BNRF1 localization to PML-nuclear bodies typically associated with host-antiviral resistance and transcriptional repression. Paradoxically, the interface is also required for selective transcription activation of viral latent cycle genes required for driving B-cell proliferation. These findings reveal molecular details of virus reprogramming of an antiviral histone chaperone to promote viral latency and cellular immortalization.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.