Impacts of DNA methylation on H2A.Z deposition and nucleosome stability
Shih, R.M., Arimura, Y., Konishi, H.A., Funabiki, H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H3.2 | 136 | Homo sapiens | Mutation(s): 0 Gene Names: H3C15, HIST2H3A, H3C14, H3F2, H3FM, HIST2H3C, H3C13, HIST2H3D | 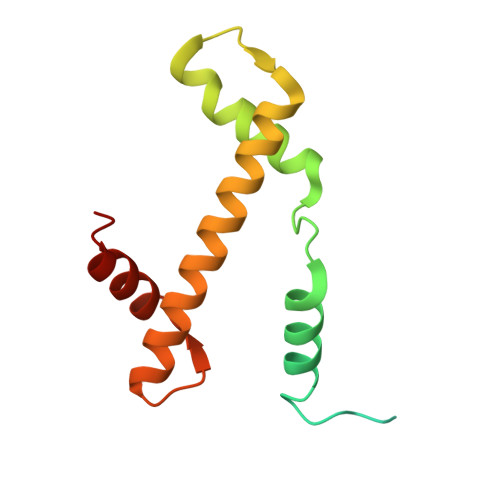 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q71DI3 (Homo sapiens) Explore Q71DI3 Go to UniProtKB: Q71DI3 | |||||
PHAROS: Q71DI3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q71DI3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H4 | 103 | Homo sapiens | Mutation(s): 0 Gene Names: | 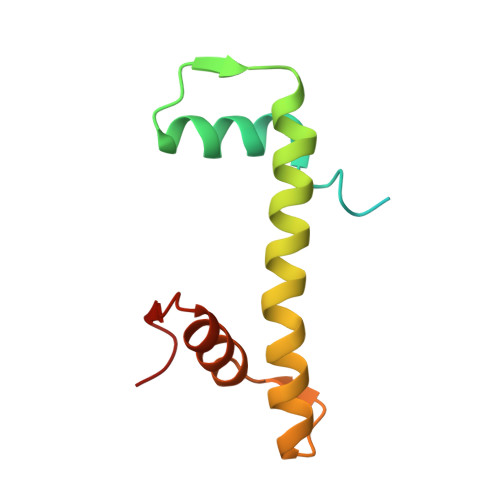 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62805 (Homo sapiens) Explore P62805 Go to UniProtKB: P62805 | |||||
PHAROS: P62805 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62805 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A.Z | 127 | Homo sapiens | Mutation(s): 0 Gene Names: H2AZ1, H2AFZ, H2AZ | 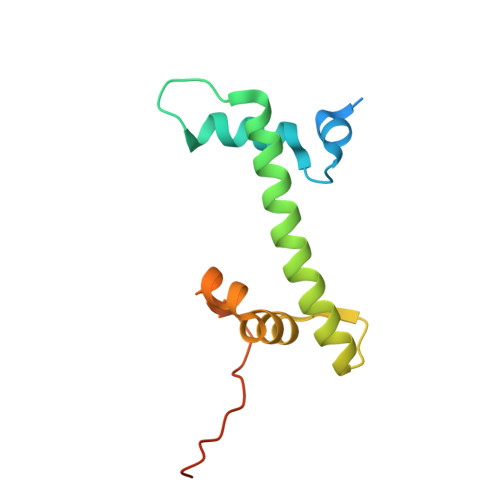 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0C0S5 (Homo sapiens) Explore P0C0S5 Go to UniProtKB: P0C0S5 | |||||
PHAROS: P0C0S5 GTEx: ENSG00000164032 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0S5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2B type 1-J | 126 | Homo sapiens | Mutation(s): 0 Gene Names: HIST1H2BJ, H2BFR | 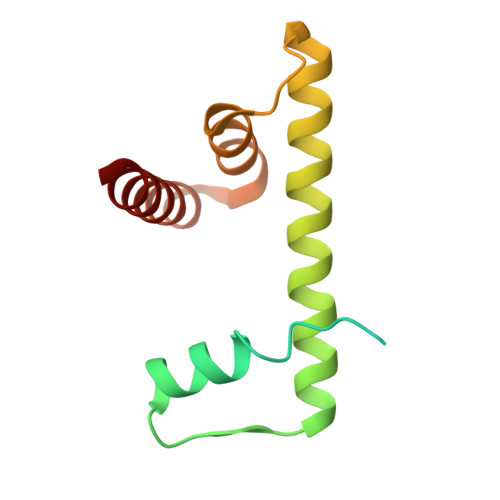 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06899 (Homo sapiens) Explore P06899 Go to UniProtKB: P06899 | |||||
PHAROS: P06899 GTEx: ENSG00000124635 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 601L DNA (137-MER) | 145 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 601L DNA (137-MER) | 145 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21_5207 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R35GM132111 |