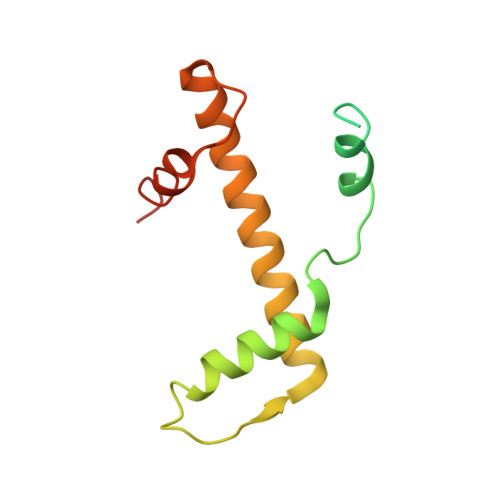
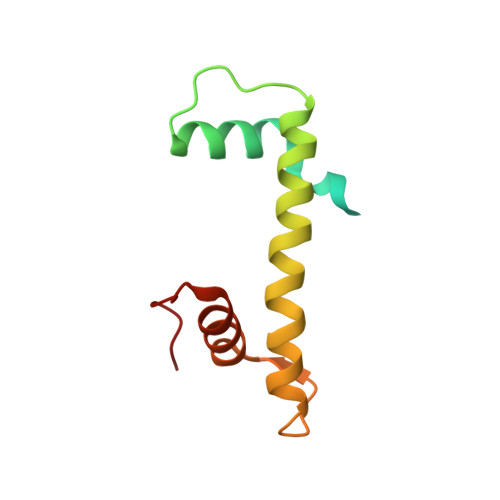
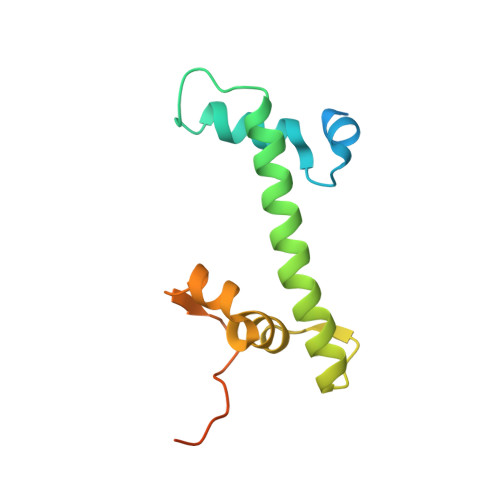
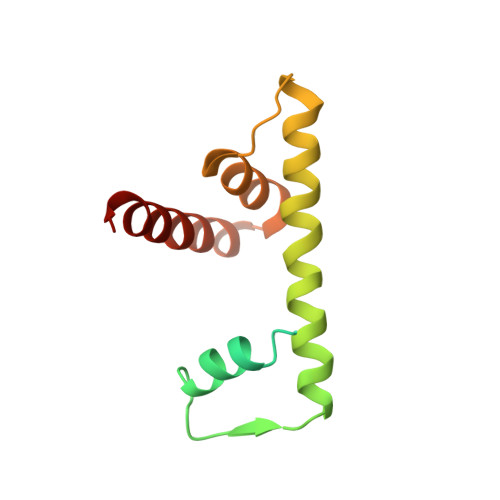
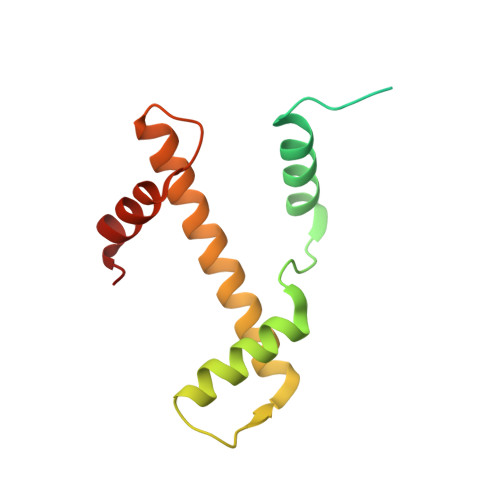
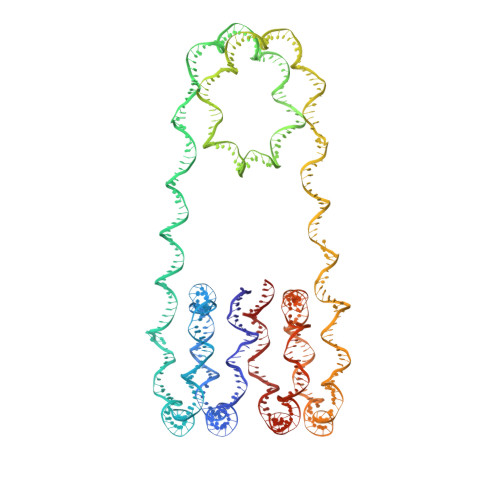
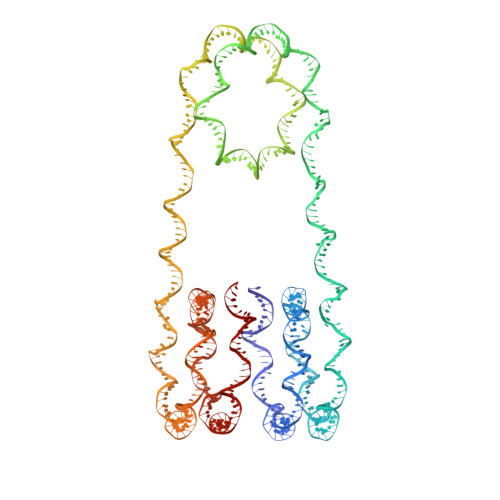
Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Takizawa, Y., Ho, C.H., Tachiwana, H., Matsunami, H., Kobayashi, W., Suzuki, M., Arimura, Y., Hori, T., Fukagawa, T., Ohi, M.D., Wolf, M., Kurumizaka, H.(2020) Structure 28: 44
- PubMed: 31711756
- DOI: https://doi.org/10.1016/j.str.2019.10.016
- Primary Citation of Related Structures:
6L49, 6L4A - PubMed Abstract:
The histone H3 variant CENP-A is a crucial epigenetic marker for centromere specification. CENP-A forms a characteristic nucleosome and dictates the higher-order configuration of centromeric chromatin. However, little is known about how the CENP-A nucleosome affects the architecture of centromeric chromatin. In this study, we reconstituted tri-nucleosomes mimicking a centromeric nucleosome arrangement containing the CENP-A nucleosome, and determined their 3D structures by cryoelectron microscopy. The H3-CENP-A-H3 tri-nucleosomes adopt an untwisted architecture, with an outward-facing linker DNA path between nucleosomes. This is distinct from the H3-H3-H3 tri-nucleosome architecture, with an inward-facing DNA path. Intriguingly, the untwisted architecture may allow the CENP-A nucleosome to be exposed to the solvent in the condensed chromatin model. These results provide a structural basis for understanding the 3D configuration of CENP-A-containing chromatin, and may explain how centromeric proteins can specifically target the CENP-A nucleosomes buried in robust amounts of H3 nucleosomes in centromeres.
- Laboratory of Chromatin Structure and Function, Institute for Quantitative Biosciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan; Molecular Cryo-Electron Microscopy Unit, Okinawa Institute of Science and Technology Graduate University, 1919-1 Tancha, Onna-son, Kunigami-gun, Okinawa 904-0495, Japan.
Organizational Affiliation: