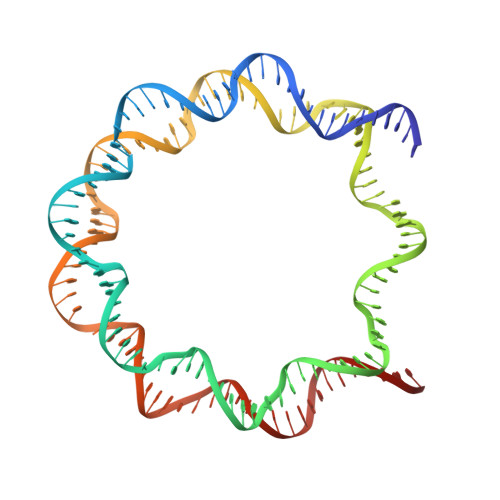
Polymorphism of apyrimidinic DNA structures in the nucleosome
Osakabe, A., Arimura, Y., Matsumoto, S., Horikoshi, N., Sugasawa, K., Kurumizaka, H.(2017) Sci Rep 7: 41783-41783
- PubMed: 28139742
- DOI: https://doi.org/10.1038/srep41783
- Primary Citation of Related Structures:
5JRG - PubMed Abstract:
Huge amounts (>10,000/day) of apurinic/apyrimidinic (AP) sites are produced in genomes, but their structures in chromatin remain undetermined. We determined the crystal structure of the nucleosome containing AP-site analogs at two symmetric sites, which revealed structural polymorphism: one forms an inchworm configuration without an empty space at the AP site, and the other forms a B-form-like structure with an empty space and the orphan base. This unexpected inchworm configuration of the AP site is important to understand the AP DNA repair mechanism, because it may not be recognized by the major AP-binding protein, APE1, during the base excision repair process.
- Laboratory of Structural Biology, Graduate School of Advanced Science and Engineering, Waseda University, 2-2 Wakamatsu-cho, Shinjuku-ku, Tokyo 162-8480, Japan.
Organizational Affiliation: