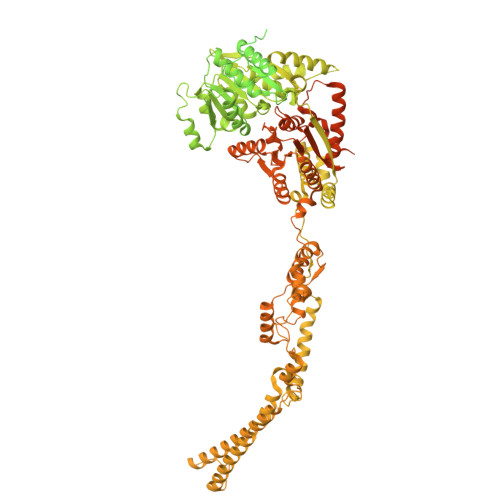
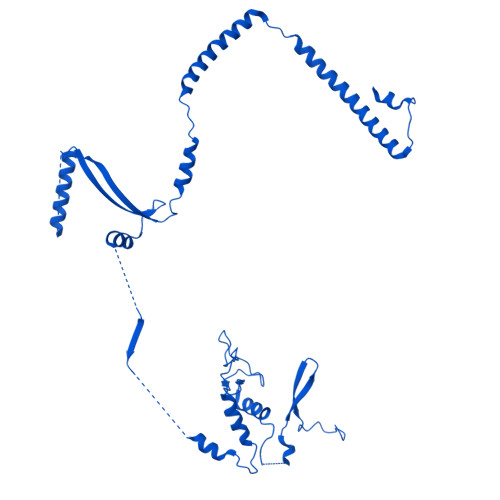
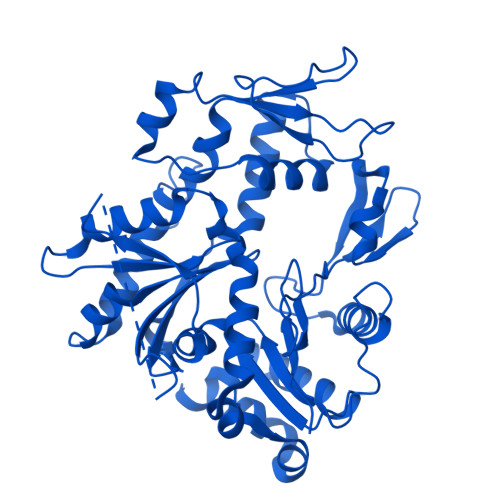
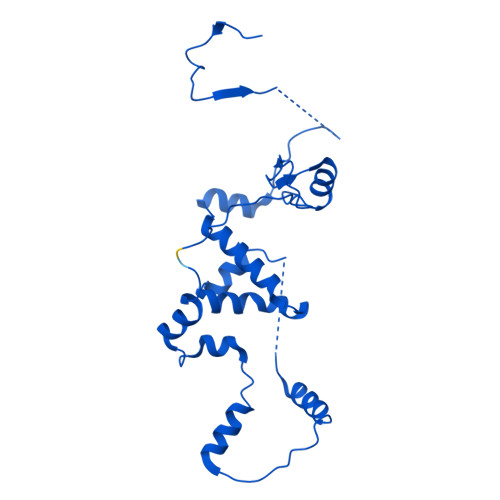
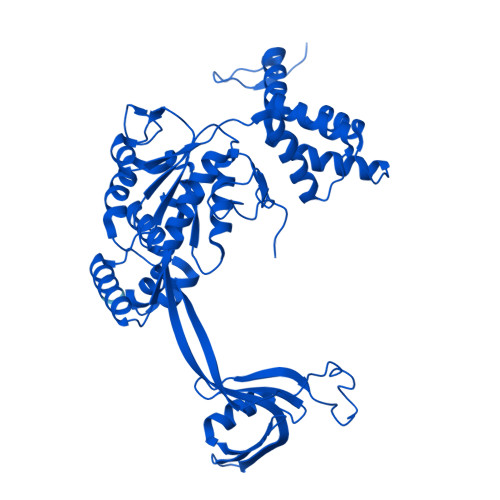
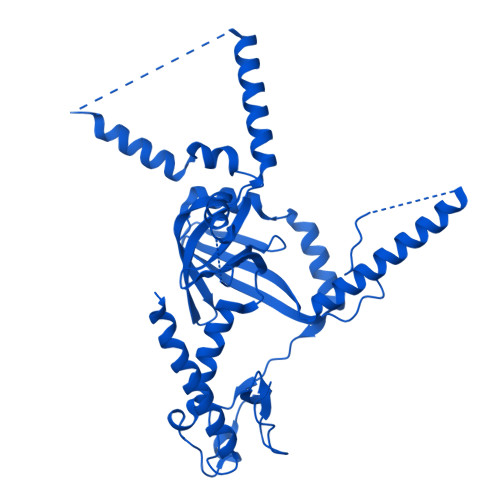
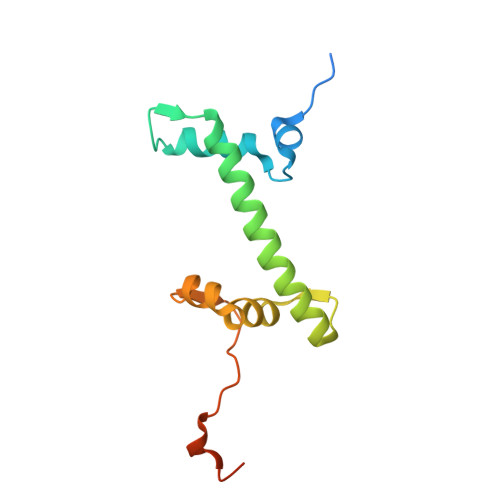
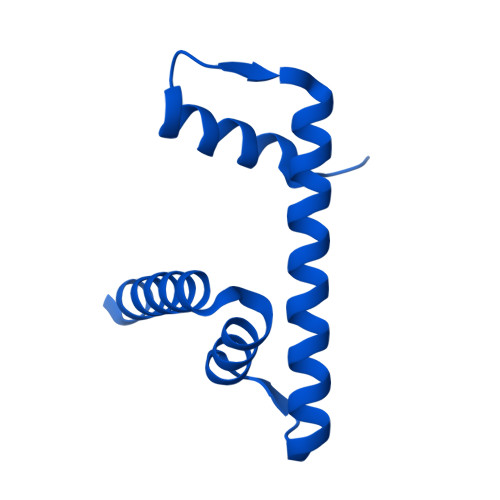
Molecular basis of global promoter sensing and nucleosome capture by the SWR1 chromatin remodeler.
Louder, R.K., Park, G., Ye, Z., Cha, J.S., Gardner, A.M., Lei, Q., Ranjan, A., Hollmuller, E., Stengel, F., Pugh, B.F., Wu, C.(2024) Cell
- PubMed: 39357520
- DOI: https://doi.org/10.1016/j.cell.2024.09.007
- Primary Citation of Related Structures:
9B1D, 9B1E - PubMed Abstract:
The SWR1 chromatin remodeling complex is recruited to +1 nucleosomes downstream of transcription start sites of eukaryotic promoters, where it exchanges histone H2A for the specialized variant H2A.Z. Here, we use cryoelectron microscopy (cryo-EM) to resolve the structural basis of the SWR1 interaction with free DNA, revealing a distinct open conformation of the Swr1 ATPase that enables sliding from accessible DNA to nucleosomes. A complete structural model of the SWR1-nucleosome complex illustrates critical roles for Swc2 and Swc3 subunits in oriented nucleosome engagement by SWR1. Moreover, an extended DNA-binding α helix within the Swc3 subunit enables sensing of nucleosome linker length and is essential for SWR1-promoter-specific recruitment and activity. The previously unresolved N-SWR1 subcomplex forms a flexible extended structure, enabling multivalent recognition of acetylated histone tails by reader domains to further direct SWR1 toward the +1 nucleosome. Altogether, our findings provide a generalizable mechanism for promoter-specific targeting of chromatin and transcription complexes.
Organizational Affiliation:
Department of Biology, Johns Hopkins University, Baltimore, MD, USA. Electronic address: rklouder@jhu.edu.