Structure of in situ PBS-PSII supercomplex at 3.5 Angstroms resolution.
You, X., Zhang, X., Xiao, Y.N., Sun, S., Sui, S.F.To be published.
Include CSM | ||
| Advanced Search | Browse Annotations | ||
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
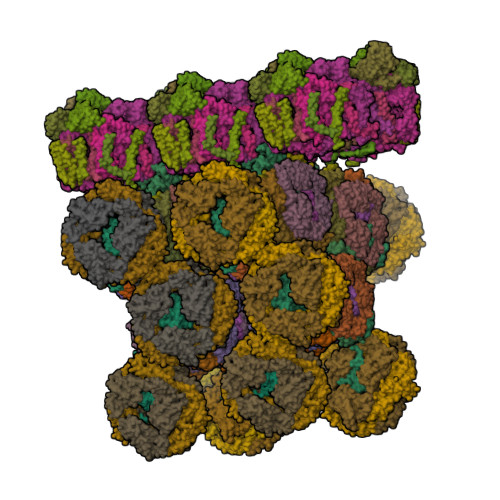
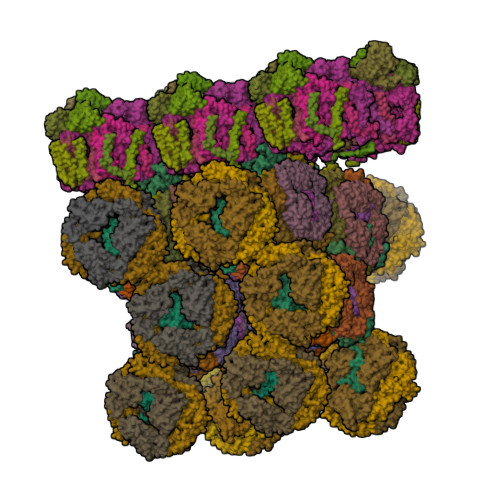
Biological assembly 1 assigned by authors and generated by (software)
Macromolecule Content
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LRC1 | 254 | Arthrospira sp. FACHB-439 | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LCM | B [auth BB], C [auth CB], EH [auth B4], FH [auth C4] | 914 | Arthrospira sp. FACHB-439 | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phycobilisome 7.8 kDa linker polypeptide, allophycocyanin-associated, core | 67 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: apcC | 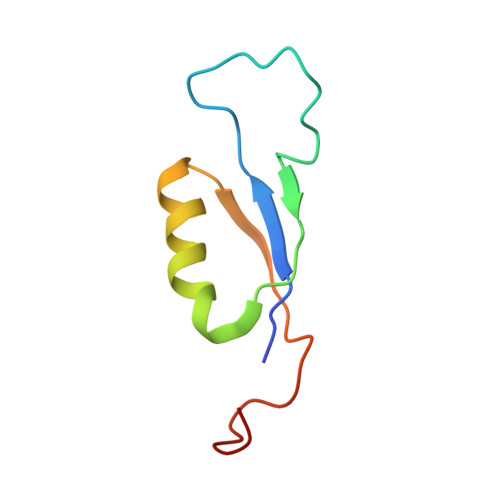 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II protein D1 | 359 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbA1 EC: 1.10.3.9 | 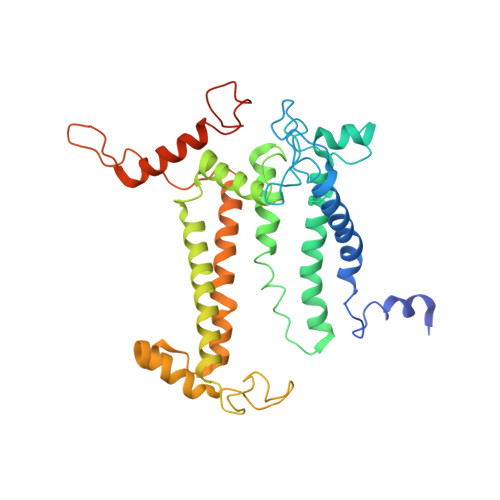 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP47 reaction center protein | 509 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbB | 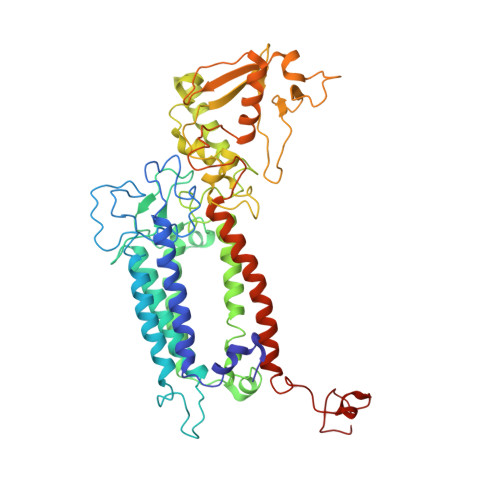 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP43 reaction center protein | 469 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbC | 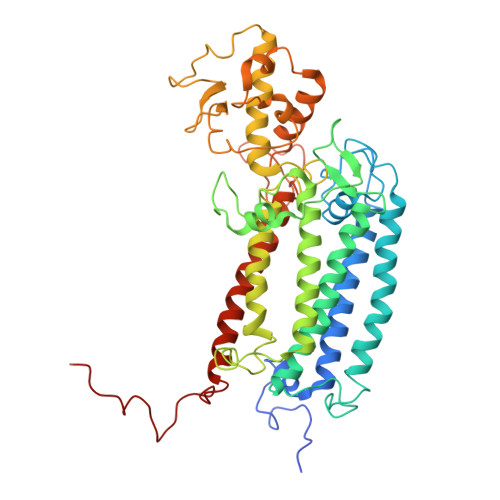 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II D2 protein | 352 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbD1, psbD EC: 1.10.3.9 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit alpha | 82 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbE |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit beta | 44 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbF |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein H | 67 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbH |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein I | 38 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbI |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein J | 39 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbJ |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein K | 45 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbK |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein L | 39 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbL |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein M | 37 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbM |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II extrinsic protein O | 278 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbO |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PsbQ protein | 164 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbQ |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Y | 39 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbY |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 21 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LPP1 | BB [auth SE], BF [auth S1], FA [auth SD] | 47 | Arthrospira sp. FACHB-439 | Mutation(s): 0 |  |
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 22 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein T | 31 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbTc, psbT |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 23 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PsbU | 95 | Arthrospira sp. FACHB-439 | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 24 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II extrinsic protein V | 163 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbV1, psbV |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 25 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein X | 48 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbX |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 26 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Psb30 | 43 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psb30 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 27 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Z | 63 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: psbZ |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 28 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LPP2 | IB [auth EF], OC [auth EK], TB [auth BG], ZC [auth BL] | 150 | Arthrospira sp. FACHB-439 | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 29 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Allophycocyanin beta-18 subunit | EC [auth QG], GN [auth gF], KD [auth QL], NP [auth gK] | 169 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: apcF |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
Entity ID: 30 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Allophycocyanin-B alpha subunit | JC [auth VG], MN [auth mF], PD [auth VL], TP [auth mK] | 161 | Arthrospira sp. FACHB-439 | Mutation(s): 0 Gene Names: apcD |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
LOADING | |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |
RCSB PDB Core Operations are funded by the U.S. National Science Foundation (DBI-2321666), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM157729.