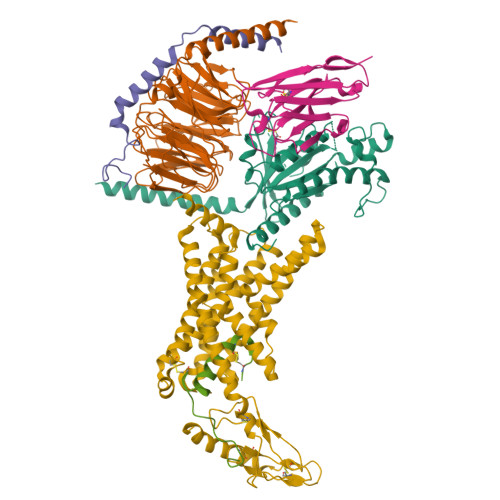
A structural basis for amylin receptor phenotype.
Cao, J., Belousoff, M.J., Liang, Y.L., Johnson, R.M., Josephs, T.M., Fletcher, M.M., Christopoulos, A., Hay, D.L., Danev, R., Wootten, D., Sexton, P.M.(2022) Science 375: eabm9609-eabm9609
- PubMed: 35324283
- DOI: https://doi.org/10.1126/science.abm9609
- Primary Citation of Related Structures:
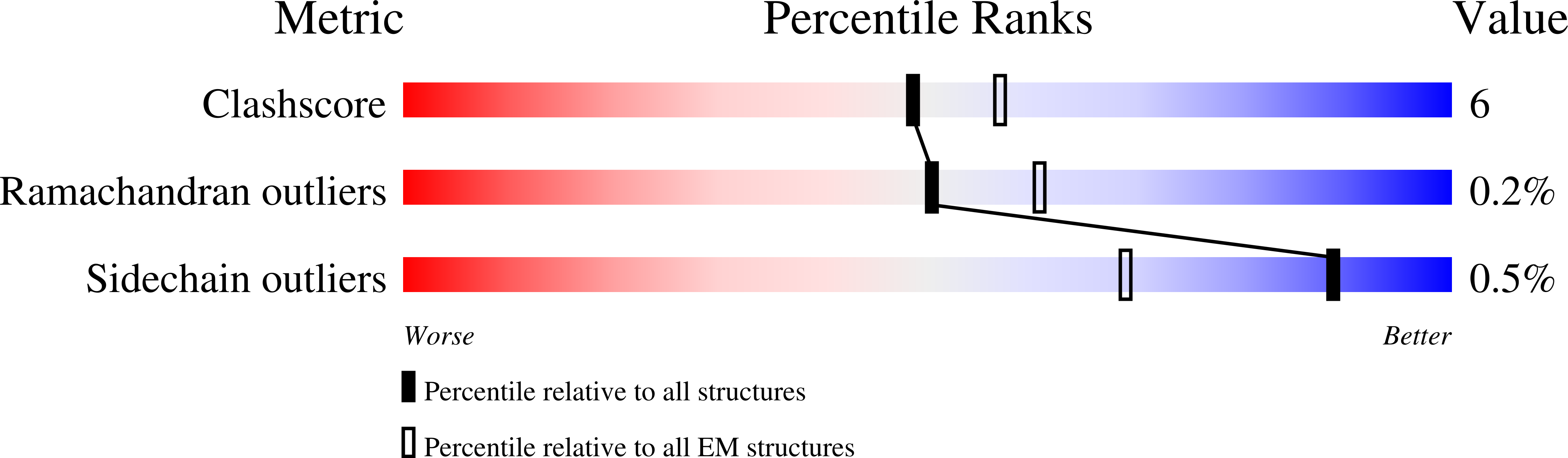
7TYF, 7TYH, 7TYI, 7TYL, 7TYN, 7TYO, 7TYW, 7TYX, 7TYY, 7TZF - PubMed Abstract:
Amylin receptors (AMYRs) are heterodimers of the calcitonin (CT) receptor (CTR) and one of three receptor activity-modifying proteins (RAMPs), AMY 1 R, AMY 2 R, and AMY 3 R. Selective AMYR agonists and dual AMYR/CTR agonists are being developed as obesity treatments; however, the molecular basis for peptide binding and selectivity is unknown. We determined the structure and dynamics of active AMYRs with amylin, AMY 1 R with salmon CT (sCT), AMY 2 R with sCT or human CT (hCT), and CTR with amylin, sCT, or hCT. The conformation of amylin-bound complexes was similar for all AMYRs, constrained by the RAMP, and an ordered midpeptide motif that we call the bypass motif. The CT-bound AMYR complexes were distinct, overlapping the CT-bound CTR complexes. Our findings indicate that activation of AMYRs by CT-based peptides is distinct from their activation by amylin-based peptides. This has important implications for the development of AMYR therapeutics.
Organizational Affiliation:
Drug Discovery Biology Theme, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, Victoria, Australia.