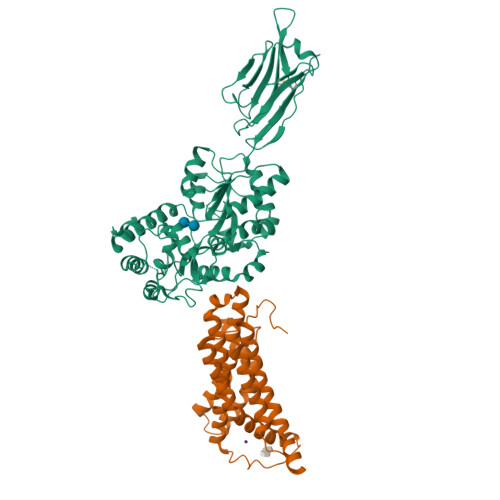
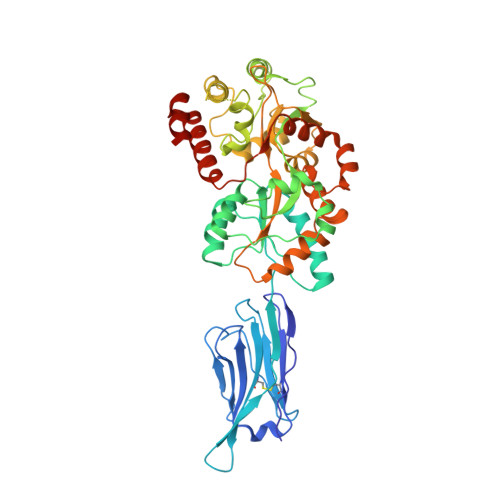
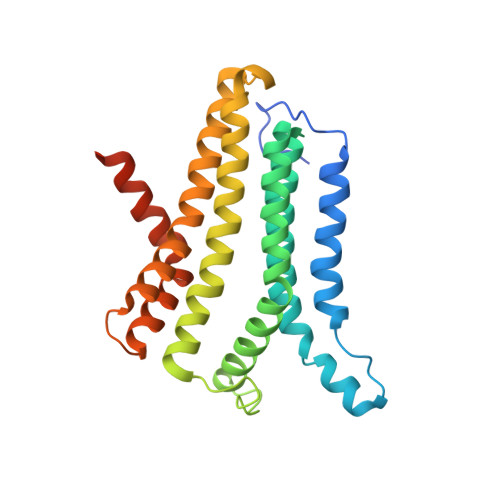
Structural basis for ion selectivity in TMEM175 K+channels.
Brunner, J.D., Jakob, R.P., Schulze, T., Neldner, Y., Moroni, A., Thiel, G., Maier, T., Schenck, S.(2020) Elife 9
- PubMed: 32267231
- DOI: https://doi.org/10.7554/eLife.53683
- Primary Citation of Related Structures:
6HD8, 6HD9, 6HDA, 6HDB, 6HDC, 6SWR - PubMed Abstract:
The TMEM175 family constitutes recently discovered K + channels that are important for autophagosome turnover and lysosomal pH regulation and are associated with the early onset of Parkinson Disease. TMEM175 channels lack a P-loop selectivity filter, a hallmark of all known K + channels, raising the question how selectivity is achieved. Here, we report the X-ray structure of a closed bacterial TMEM175 channel in complex with a nanobody fusion-protein disclosing bound K + ions. Our analysis revealed that a highly conserved layer of threonine residues in the pore conveys a basal K + selectivity. An additional layer comprising two serines in human TMEM175 increases selectivity further and renders this channel sensitive to 4-aminopyridine and Zn 2+ . Our findings suggest that large hydrophobic side chains occlude the pore, forming a physical gate, and that channel opening by iris-like motions simultaneously relocates the gate and exposes the otherwise concealed selectivity filter to the pore lumen.
Organizational Affiliation:
Department of Biochemistry, University of Zürich, Zürich, Switzerland.