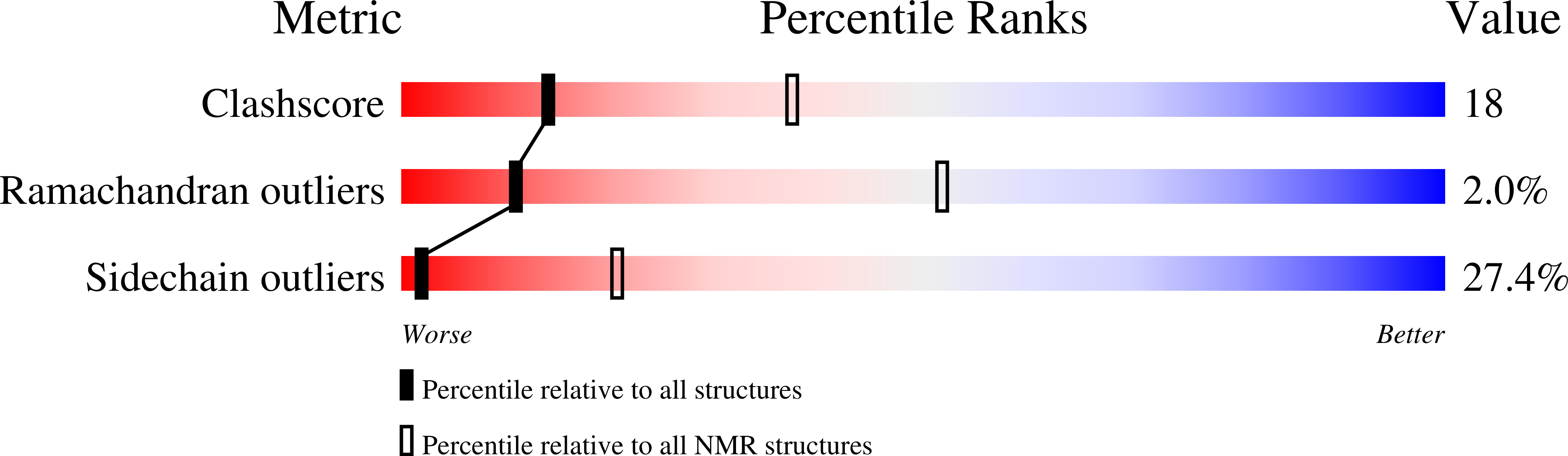Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
Eichmann, C., Tzitzilonis, C., Bordignon, E., Maslennikov, I., Choe, S., Riek, R.(2014) J Biological Chem 289: 23482-23503
- PubMed: 24958726
- DOI: https://doi.org/10.1074/jbc.M114.571935
- Primary Citation of Related Structures:
2MOI, 2MOL, 2MPN - PubMed Abstract:
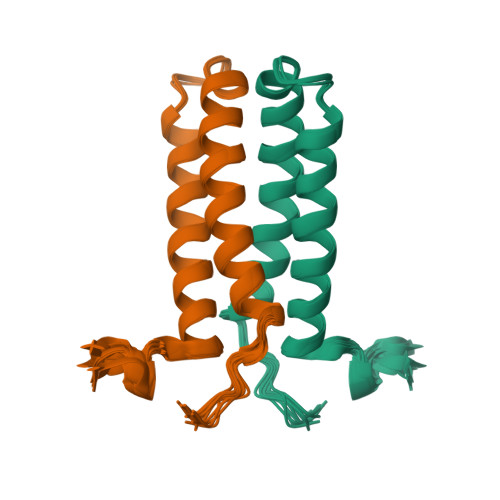
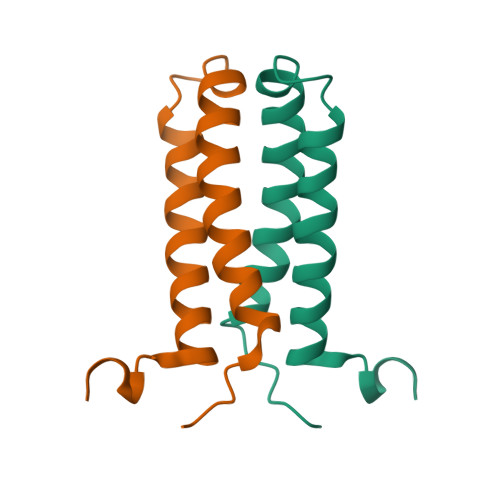
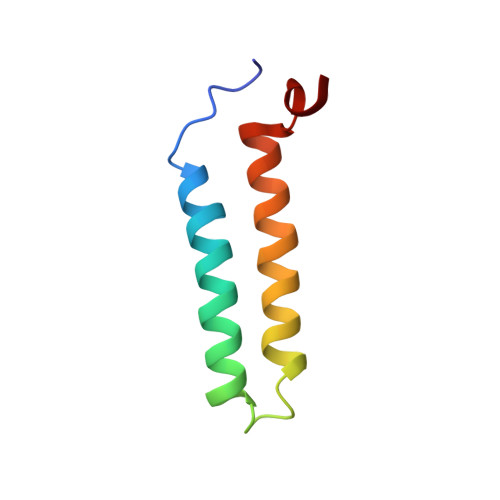
The solution NMR structure of the α-helical integral membrane protein YgaP from Escherichia coli in mixed 1,2-diheptanoyl-sn-glycerol-3-phosphocholine/1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1'-rac-glycerol) micelles is presented. In these micelles, YgaP forms a homodimer with the two transmembrane helices being the dimer interface, whereas the N-terminal cytoplasmic domain includes a rhodanese-fold in accordance to its sequence homology to the rhodanese family of sulfurtransferases. The enzymatic sulfur transfer activity of full-length YgaP as well as of the N-terminal rhodanese domain only was investigated performing a series of titrations with sodium thiosulfate and potassium cyanide monitored by NMR and EPR. The data indicate the thiosulfate concentration-dependent addition of several sulfur atoms to the catalytic Cys-63, which process can be reversed by the addition of potassium cyanide. The catalytic reaction induces thereby conformational changes within the rhodanese domain, as well as on the transmembrane α-helices of YgaP. These results provide insights into a potential mechanism of YgaP during the catalytic thiosulfate activity in vivo.
Organizational Affiliation:
From the Laboratory of Physical Chemistry, Swiss Federal Institute of Technology, ETH-Hönggerberg, CH-8093 Zürich, Switzerland and.