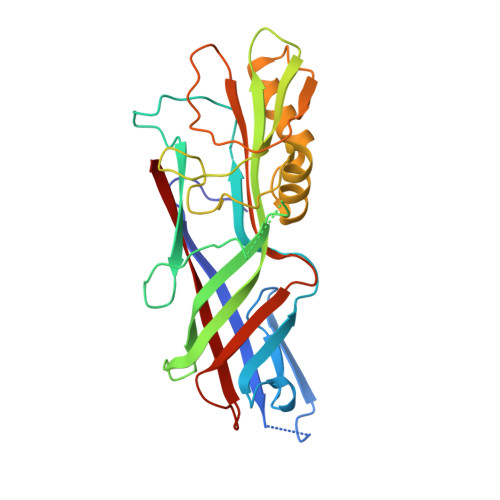
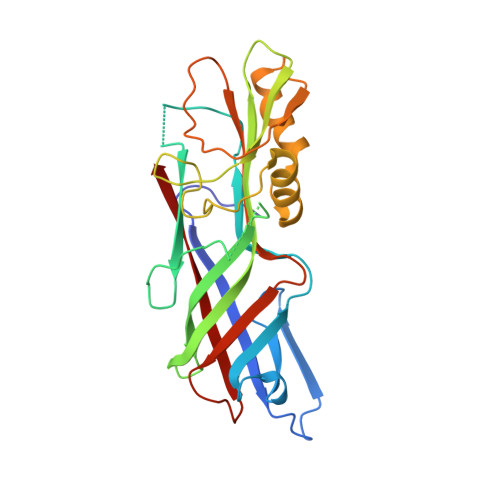
Chloroplasts Assemble the Major Subunit Faeg of Escherichia Coli F4 (K88) Fimbriae Into Strand-Swapped Dimers
Van Molle, I., Joensuu, J.J., Buts, L., Panjikar, S., Kotiaho, M., Bouckaert, J., Wyns, L., Niklander-Teeri, V., De Greve, H.(2007) J Mol Biol 368: 791
- PubMed: 17368480
- DOI: https://doi.org/10.1016/j.jmb.2007.02.051
- Primary Citation of Related Structures:
2J6G, 2J6R - PubMed Abstract:
F4 fimbriae encoded by the fae operon are the major colonization factors associated with porcine neonatal and postweaning diarrhoea caused by enterotoxigenic Escherichia coli (ETEC). Via the chaperone/usher pathway, the F4 fimbriae are assembled as long polymers of the major subunit FaeG, which also possesses the adhesive properties of the fimbriae. Intrinsically, the incomplete fold of fimbrial subunits renders them unstable and susceptible to aggregation and/or proteolytic degradation in the absence of a specific periplasmic chaperone. In order to test the possibility of producing FaeG in plants, FaeG expression was studied in transgenic tobacco plants. FaeG was directed to different subcellular compartments by specific targeting signals. Targeting of FaeG to the chloroplast results in much higher yields than FaeG targeting to the endoplasmic reticulum or the apoplast. Two chloroplast-targeted FaeG variants were purified from tobacco plants and crystallized. The crystal structures show that chloroplasts circumvent the absence of the fimbrial assembly machinery by assembling FaeG into strand-swapped dimers. Furthermore, the structures reveal how FaeG combines the structural requirements of a major fimbrial subunit with its adhesive role by grafting an additional domain on its Ig-like core.
Organizational Affiliation:
Department of Molecular and Cellular Interactions, Flanders Institute for Biotechnology (VIB), Brussels, Belgium.