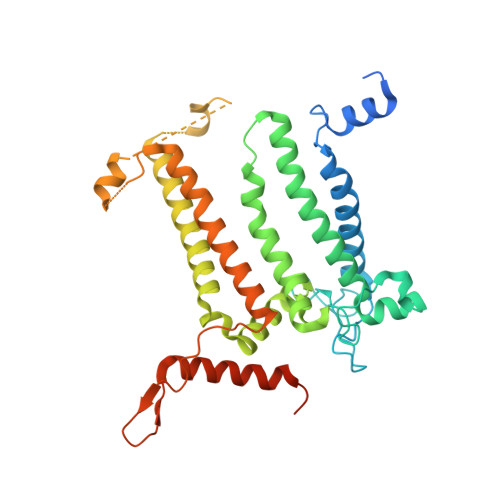
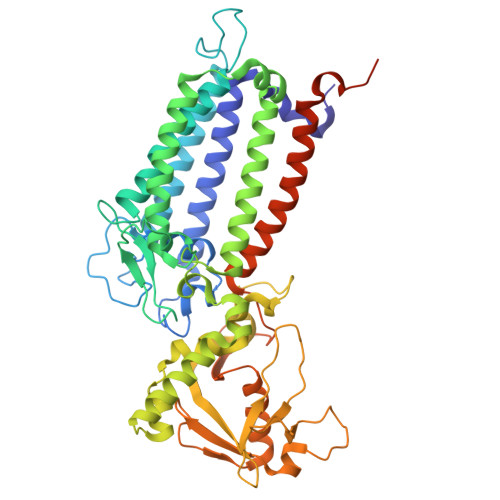
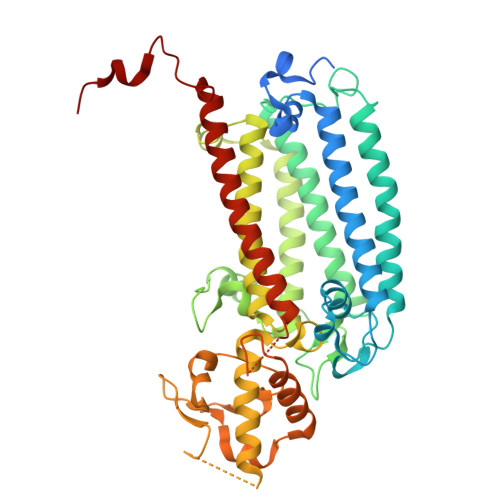
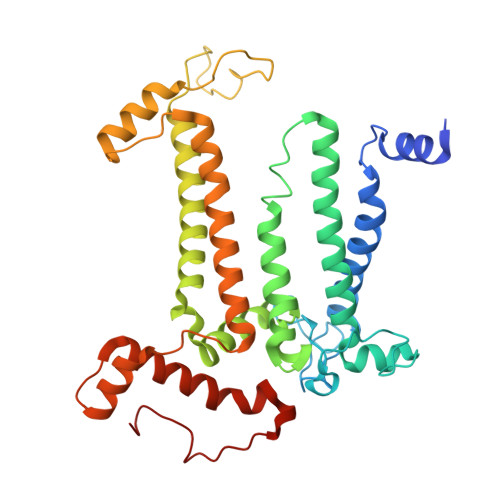
Structural analysis of PSI-ACPI and PSII-ACPII supercomplexes from a cryptophyte alga Rhodomonas sp. NIES-2332.
Zhang, W., Yonehara, N., Ishii, M., Jiang, H., La Rocca, R., Tsai, P.C., Li, H., Kato, K., Akita, F., Shen, J.R.(2025) Front Plant Sci 16: 1716939-1716939
- PubMed: 41393888
- DOI: https://doi.org/10.3389/fpls.2025.1716939
- Primary Citation of Related Structures:
9KZ9, 9L0K, 9L5V - PubMed Abstract:
Light energy is converted to chemical energy by two photosystems (PSI and PSII) in complex with their light-harvesting complex proteins (LHCI and LHCII) in photosynthesis. Rhodomonas is a member of cryptophyte alga whose LHCs contain unique chlorophyll a/c proteins (ACPs) and phycobiliproteins. We purified PSI-ACPI and PSII-ACPII supercomplexes from a cryptophyte Rhodomonas sp. NIES-2332 and analyzed their structures at high resolutions of 2.08 Å and 2.17 Å, respectively, using cryo-electron microscopy. These structures are largely similar to those reported previously from two other species of cryptophytes, but exhibited some differences in both the pigment locations and subunit structures. A part of the antenna subunits of both photosystems is shifted compared with the previously reported structures from other species of cryptophytes, suggesting some differences in the energy transfer rates from the antenna to the PSI and PSII cores. Newly identified lipids are found to occupy the interfaces between the antennae and cores, which may be important for assembly and stabilization of the supercomplexes. Water molecules surrounding three iron-sulfur clusters of the PSI core are found in our high-resolution structure, some of which are conserved from cyanobacteria to higher plants but some are different. In addition, our structure of PSII-ACPII lacks the subunits of oxygen-evolving complex as well as the Mn 4 CaO 5 cluster, suggesting that the cells are in the S-growth phase, yet the PSI-ACPI structure showed the binding of PsaQ, suggesting that it is in an L-phase. These results suggest that the S-phase and L-phase can co-exist in the cryptophytic cells. The high-resolution structures of both PSI-ACPIs and PSII-ACPIIs solved in this study provide a more solid structural basis for elucidating the energy transfer and quenching mechanisms in this group of the organisms.
- Advanced Research Field, Research Institute for Interdisciplinary Science, and Graduate School of Environmental, Life, Natural Science and Technology, Okayama University, Okayama, Japan.
Organizational Affiliation: