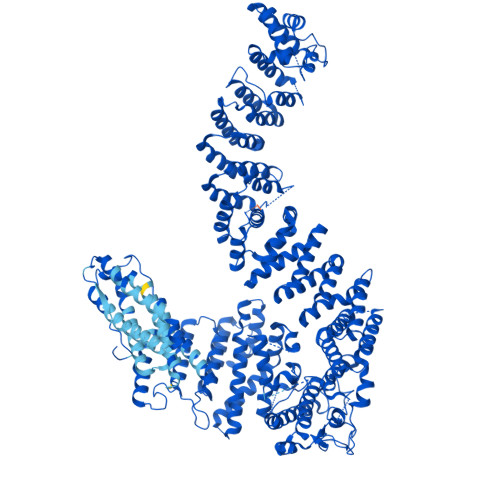
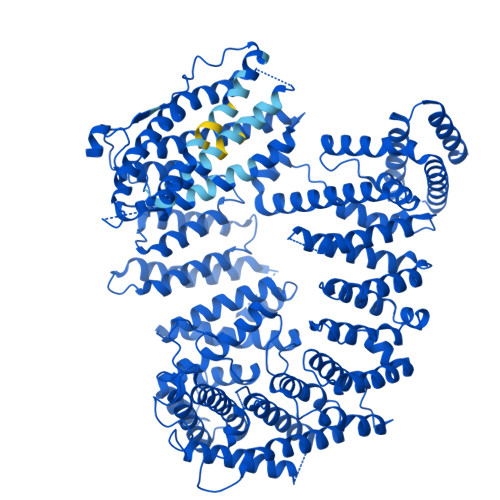
FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Alcon, P., Kaczmarczyk, A.P., Ray, K.K., Liolios, T., Guilbaud, G., Sijacki, T., Shen, Y., McLaughlin, S.H., Sale, J.E., Knipscheer, P., Rueda, D.S., Passmore, L.A.(2024) Nature 632: 1165-1173
- PubMed: 39085614
- DOI: https://doi.org/10.1038/s41586-024-07770-w
- Primary Citation of Related Structures:
9FFB, 9FFF - PubMed Abstract:
DNA crosslinks block DNA replication and are repaired by the Fanconi anaemia pathway. The FANCD2-FANCI (D2-I) protein complex is central to this process as it initiates repair by coordinating DNA incisions around the lesion 1 . However, D2-I is also known to have a more general role in DNA repair and in protecting stalled replication forks from unscheduled degradation 2-4 . At present, it is unclear how DNA crosslinks are recognized and how D2-I functions in replication fork protection. Here, using single-molecule imaging, we show that D2-I is a sliding clamp that binds to and diffuses on double-stranded DNA. Notably, sliding D2-I stalls on encountering single-stranded-double-stranded (ss-ds) DNA junctions, structures that are generated when replication forks stall at DNA lesions 5 . Using cryogenic electron microscopy, we determined structures of D2-I on DNA that show that stalled D2-I makes specific interactions with the ss-dsDNA junction that are distinct from those made by sliding D2-I. Thus, D2-I surveys dsDNA and, when it reaches an ssDNA gap, it specifically clamps onto ss-dsDNA junctions. Because ss-dsDNA junctions are found at stalled replication forks, D2-I can identify sites of DNA damage. Therefore, our data provide a unified molecular mechanism that reconciles the roles of D2-I in the recognition and protection of stalled replication forks in several DNA repair pathways.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, UK.