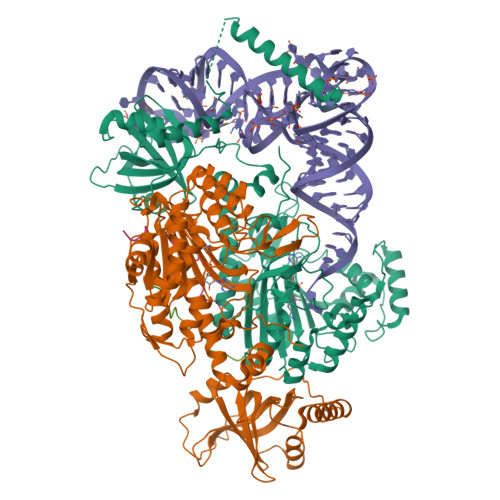
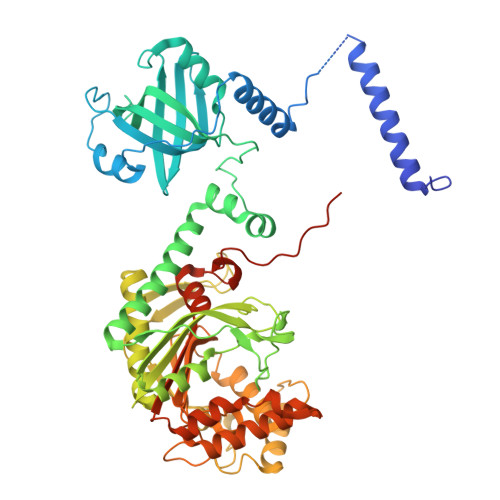
Structural basis for aminoacylation of cellular modified tRNALys3 by human lysyl-tRNA synthetase.
Devarkar, S.C., Budding, C.R., Pathirage, C., Kavoor, A., Herbert, C., Limbach, P.A., Musier-Forsyth, K., Xiong, Y.(2025) Nucleic Acids Res 53
- PubMed: 40036503
- DOI: https://doi.org/10.1093/nar/gkaf114
- Primary Citation of Related Structures:
9DOW, 9DPA, 9DPB, 9DPL - PubMed Abstract:
The average eukaryotic transfer ribonucleic acid (tRNA) contains 13 post-transcriptional modifications; however, their functional impact is largely unknown. Our understanding of the complex tRNA aminoacylation machinery in metazoans also remains limited. Herein, using a series of high-resolution cryo-electron microscopy (cryo-EM) structures, we provide the mechanistic basis for recognition and aminoacylation of fully modified cellular tRNALys3 by human lysyl-tRNA synthetase (h-LysRS). The tRNALys3 anticodon loop modifications S34 (mcm5s2U) and R37 (ms2t6A) play an integral role in recognition by h-LysRS. Modifications in the T-, variable-, and D-loops of tRNALys3 are critical for ordering the metazoan-specific N-terminal domain of LysRS. The two catalytic steps of tRNALys3 aminoacylation are structurally ordered; docking of the 3'-CCA end in the active site cannot proceed until the lysyl-adenylate intermediate is formed and the pyrophosphate byproduct is released. Association of the h-LysRS-tRNALys3 complex with a multi-tRNA synthetase complex-derived peptide shifts the equilibrium toward the 3'-CCA end "docked" conformation and allosterically increases h-LysRS catalytic efficiency. The insights presented here have broad implications for understanding the role of tRNA modifications in protein synthesis, the human aminoacylation machinery, and the growing catalog of metabolic and neurological diseases linked to it.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511, United States.