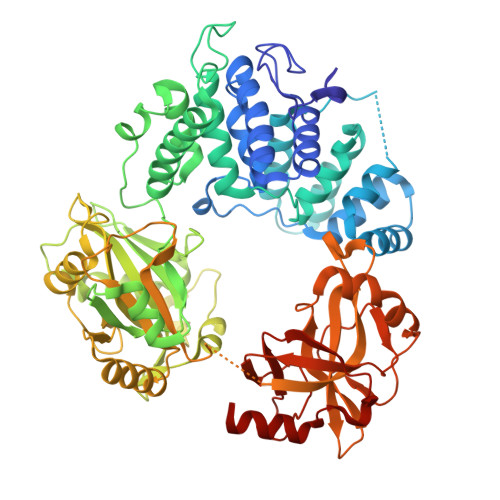
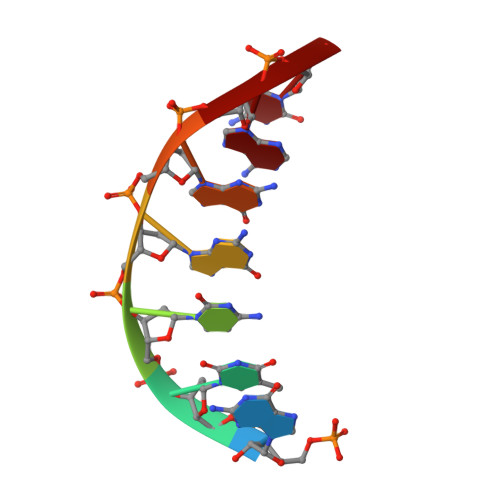
Structures of LIG1 uncover the mechanism of sugar discrimination against 5'-RNA-DNA junctions during ribonucleotide excision repair.
Balu, K.E., Tang, Q., Almohdar, D., Ratcliffe, J., Kalaycioglu, M., Caglayan, M.(2024) J Biol Chem 300: 107688-107688
- PubMed: 39159820
- DOI: https://doi.org/10.1016/j.jbc.2024.107688
- Primary Citation of Related Structures:
9BS3, 9BS4 - PubMed Abstract:
Ribonucleotides in DNA cause several types of genome instability and can be removed by ribonucleotide excision repair (RER) that is finalized by DNA ligase 1 (LIG1). However, the mechanism by which LIG1 discriminates the RER intermediate containing a 5'-RNA-DNA lesion generated by RNase H2-mediated cleavage of ribonucleotides at atomic resolution remains unknown. Here, we determine X-ray structures of LIG1/5'-rG:C at the initial step of ligation where AMP is bound to the active site of the ligase and uncover a large conformational change downstream the nick resulting in a shift at Arg(R)871 residue in the Adenylation domain of the ligase. Furthermore, we demonstrate a diminished ligation of the nick DNA substrate with a 5'-ribonucleotide in comparison to an efficient end joining of the nick substrate with a 3'-ribonucleotide by LIG1. Finally, our results demonstrate that mutations at the active site residues of the ligase and LIG1 disease-associated variants significantly impact the ligation efficiency of RNA-DNA heteroduplexes harboring "wrong" sugar at 3'- or 5'-end of nick. Collectively, our findings provide a novel atomic insight into proficient sugar discrimination by LIG1 during the processing of the most abundant form of DNA damage in cells, genomic ribonucleotides, during the initial step of the RER pathway.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Florida, Gainesville, Florida, USA.