High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesiclesCoupland, C.E., Karimi, R., Bueler, S.A., Liang, Y., Courbon, G.M., Di Trani, J.M., Wong, C.J., Saghian, R., Youn, J.Y., Wang, L.Y., Rubinstein, J.L.
(2024) Science
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) Science
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| H(+)-transporting two-sector ATPase | 646 | Rattus norvegicus | Mutation(s): 0 EC: 7.1.2.2 | 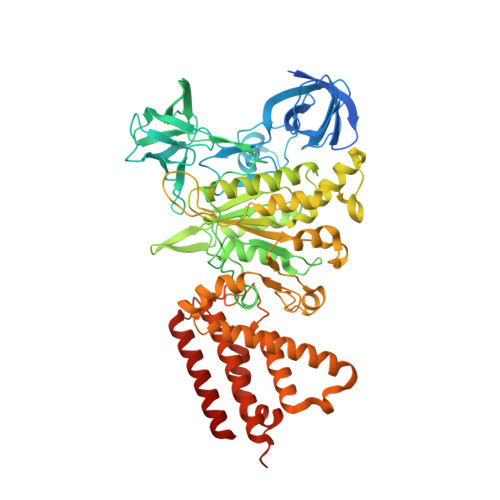 | |
UniProt | |||||
Find proteins for D4A133 (Rattus norvegicus) Explore D4A133 Go to UniProtKB: D4A133 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D4A133 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit B, brain isoform | 511 | Rattus norvegicus | Mutation(s): 0 | 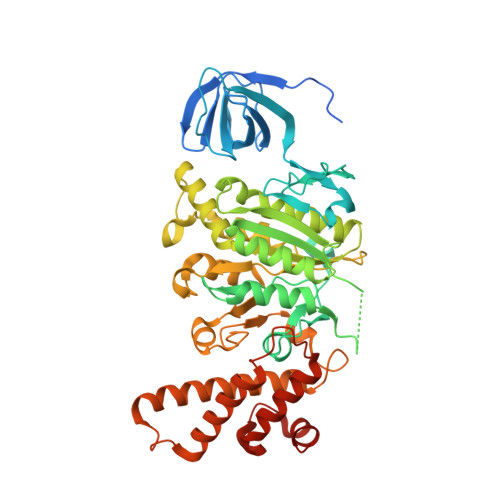 | |
UniProt | |||||
Find proteins for P62815 (Rattus norvegicus) Explore P62815 Go to UniProtKB: P62815 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62815 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit C 1 | 382 | Rattus norvegicus | Mutation(s): 0 | 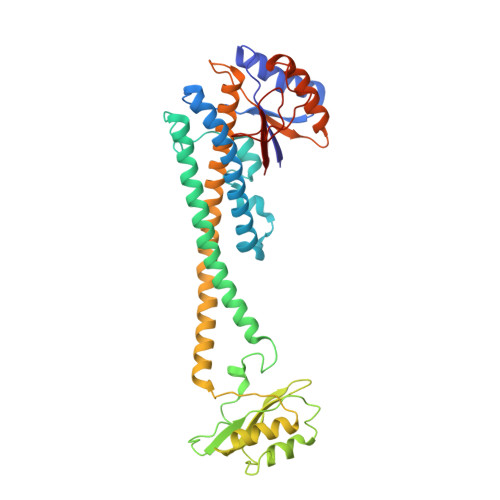 | |
UniProt | |||||
Find proteins for Q5FVI6 (Rattus norvegicus) Explore Q5FVI6 Go to UniProtKB: Q5FVI6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5FVI6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATPase H+-transporting V1 subunit D | 247 | Rattus norvegicus | Mutation(s): 0 | 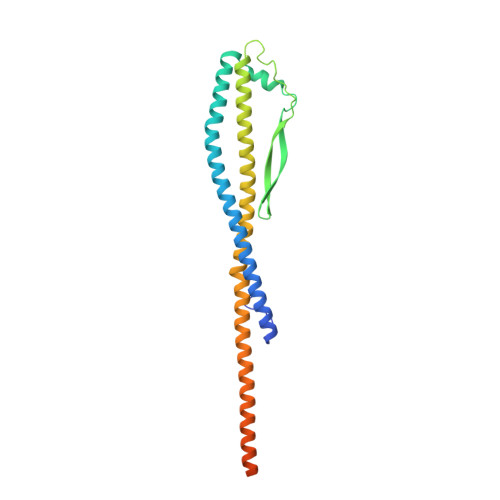 | |
UniProt | |||||
Find proteins for Q6P503 (Rattus norvegicus) Explore Q6P503 Go to UniProtKB: Q6P503 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6P503 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit E 1 | 226 | Rattus norvegicus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for Q6PCU2 (Rattus norvegicus) Explore Q6PCU2 Go to UniProtKB: Q6PCU2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6PCU2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit F | 119 | Rattus norvegicus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for P50408 (Rattus norvegicus) Explore P50408 Go to UniProtKB: P50408 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P50408 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit G | 118 | Rattus norvegicus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for Q8R2H0 (Rattus norvegicus) Explore Q8R2H0 Go to UniProtKB: Q8R2H0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8R2H0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit S1 | 463 | Rattus norvegicus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for O54715 (Rattus norvegicus) Explore O54715 Go to UniProtKB: O54715 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O54715 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Type IV secretion protein Dot | 573 | Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | Mutation(s): 0 Gene Names: lpg0968 |  | |
UniProt | |||||
Find proteins for Q5ZWW6 (Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513)) Explore Q5ZWW6 Go to UniProtKB: Q5ZWW6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ZWW6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit H | 463 | Rattus norvegicus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A8I5ZQ24 (Rattus norvegicus) Explore A0A8I5ZQ24 Go to UniProtKB: A0A8I5ZQ24 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8I5ZQ24 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Synaptophysin | 307 | Rattus norvegicus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for P07825 (Rattus norvegicus) Explore P07825 Go to UniProtKB: P07825 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07825 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase 116 kDa subunit a 1 | V [auth a] | 832 | Rattus norvegicus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P25286 (Rattus norvegicus) Explore P25286 Go to UniProtKB: P25286 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P25286 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATPase, H+ transporting, V0 subunit B (Predicted), isoform CRA_a | W [auth b] | 205 | Rattus norvegicus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for B0K022 (Rattus norvegicus) Explore B0K022 Go to UniProtKB: B0K022 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B0K022 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit | X [auth d] | 351 | Rattus norvegicus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5M7T6 (Rattus norvegicus) Explore Q5M7T6 Go to UniProtKB: Q5M7T6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5M7T6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase subunit e 2 | Y [auth e] | 81 | Rattus norvegicus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5EB76 (Rattus norvegicus) Explore Q5EB76 Go to UniProtKB: Q5EB76 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5EB76 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Rnasek protein | Z [auth f] | 86 | Rattus norvegicus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8J8YMT9 (Rattus norvegicus) Explore A0A8J8YMT9 Go to UniProtKB: A0A8J8YMT9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8J8YMT9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type proton ATPase 16 kDa proteolipid subunit | 155 | Rattus norvegicus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for P63081 (Rattus norvegicus) Explore P63081 Go to UniProtKB: P63081 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63081 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Renin receptor | JA [auth p] | 350 | Rattus norvegicus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q6AXS4 (Rattus norvegicus) Explore Q6AXS4 Go to UniProtKB: Q6AXS4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6AXS4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PC1 Query on PC1 | DB [auth b], UA [auth P], UB [auth l], VA [auth P], WA [auth a] | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE C44 H88 N O8 P NRJAVPSFFCBXDT-HUESYALOSA-N |  | ||
| PTY Query on PTY | EB [auth b] FB [auth b] LC [auth p] MC [auth p] NC [auth p] | PHOSPHATIDYLETHANOLAMINE C40 H80 N O8 P NJGIRBISCGPRPF-KXQOOQHDSA-N |  | ||
| WJP Query on WJP | AB [auth a] | methyl (3R,6Z,10E,14E)-3,7,11,15,19-pentamethylicosa-6,10,14,18-tetraen-1-yl dihydrogen diphosphate C26 H48 O7 P2 GDOCHZBDTMOKKR-FEJNUDJDSA-N |  | ||
| LP3 Query on LP3 | CB [auth a] | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE C26 H55 N O7 P IHNKQIMGVNPMTC-RUZDIDTESA-O |  | ||
| ADP Query on ADP | TA [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| CLR Query on CLR | AC [auth m] BC [auth m] CC [auth m] DC [auth n] EC [auth n] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| NAG Query on NAG | BB [auth a] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21_5207: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | PJT166152 |