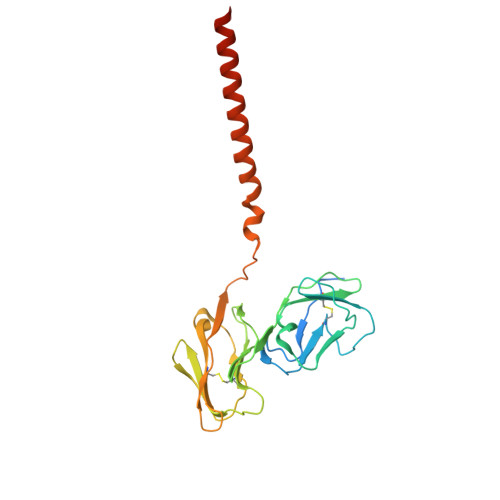
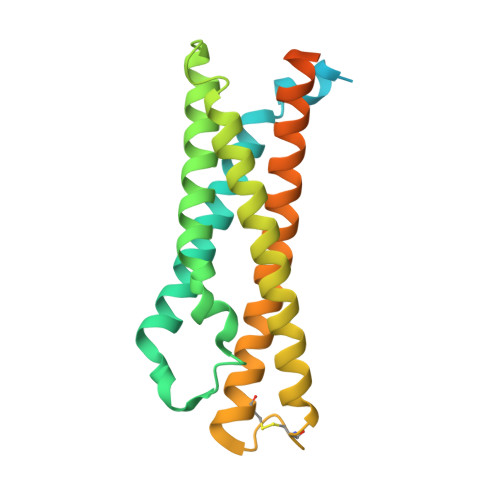
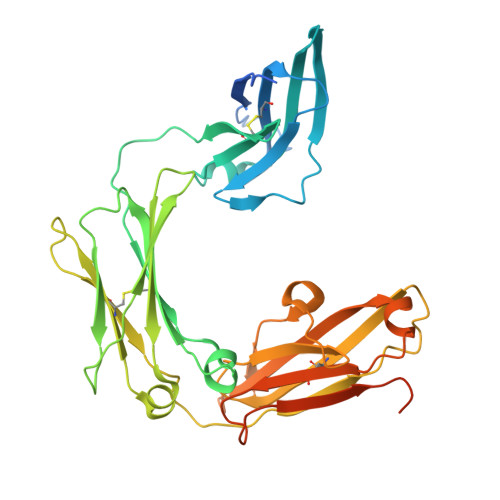
Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Deng, M., Du, S., Hou, H., Xiao, J.(2024) Nature 633: 952-959
- PubMed: 39169187
- DOI: https://doi.org/10.1038/s41586-024-07864-5
- Primary Citation of Related Structures:
8Y81, 8Y84, 8Z0T, 8ZGS, 8ZGT - PubMed Abstract:
Immunoglobulin E (IgE) plays a pivotal role in allergic responses 1,2 . The high-affinity IgE receptor, FcεRI, found on mast cells and basophils, is central to the effector functions of IgE. FcεRI is a tetrameric complex, comprising FcεRIα, FcεRIβ and a homodimer of FcRγ (originally known as FcεRIγ), with FcεRIα recognizing the Fc region of IgE (Fcε) and FcεRIβ-FcRγ facilitating signal transduction 3 . Additionally, FcRγ is a crucial component of other immunoglobulin receptors, including those for IgG (FcγRI and FcγRIIIA) and IgA (FcαRI) 4-8 . However, the molecular basis of FcεRI assembly and the structure of FcRγ have remained elusive. Here we elucidate the cryogenic electron microscopy structure of the Fcε-FcεRI complex. FcεRIα has an essential role in the receptor's assembly, interacting with FcεRIβ and both FcRγ subunits. FcεRIβ is structured as a compact four-helix bundle, similar to the B cell antigen CD20. The FcRγ dimer exhibits an asymmetric architecture, and coils with the transmembrane region of FcεRIα to form a three-helix bundle. A cholesterol-like molecule enhances the interaction between FcεRIβ and the FcεRIα-FcRγ complex. Our mutagenesis analyses further indicate similarities between the interaction of FcRγ with FcεRIα and FcγRIIIA, but differences in that with FcαRI. These findings deepen our understanding of the signalling mechanisms of FcεRI and offer insights into the functionality of other immune receptors dependent on FcRγ.
- State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, Beijing, People's Republic of China.
Organizational Affiliation: