Structure of the high affinity receptor fc(epsilon)ri TM
Du, S., Deng, M.J., Xiao, J.Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| High affinity immunoglobulin epsilon receptor subunit alpha | 245 | Rattus norvegicus | Mutation(s): 0 Gene Names: Fcer1a, Fce1a Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P12371 (Rattus norvegicus) Explore P12371 Go to UniProtKB: P12371 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12371 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| High affinity immunoglobulin epsilon receptor subunit beta | 243 | Rattus norvegicus | Mutation(s): 0 Gene Names: Fce1b, Fcer1b Membrane Entity: Yes | 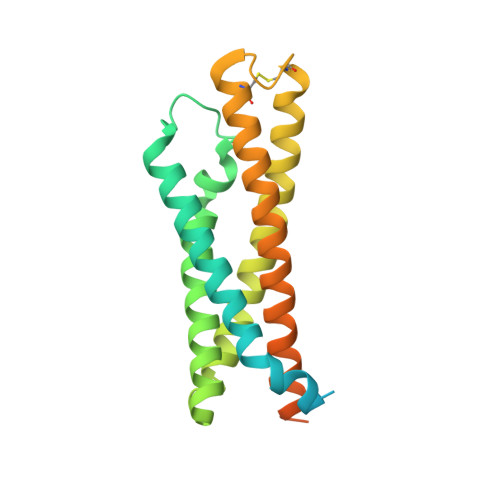 | |
UniProt | |||||
Find proteins for P13386 (Rattus norvegicus) Explore P13386 Go to UniProtKB: P13386 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13386 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| High affinity immunoglobulin epsilon receptor subunit gamma | C, D [auth G] | 119 | Rattus norvegicus | Mutation(s): 0 Gene Names: Fcer1g, Fce1g Membrane Entity: Yes | 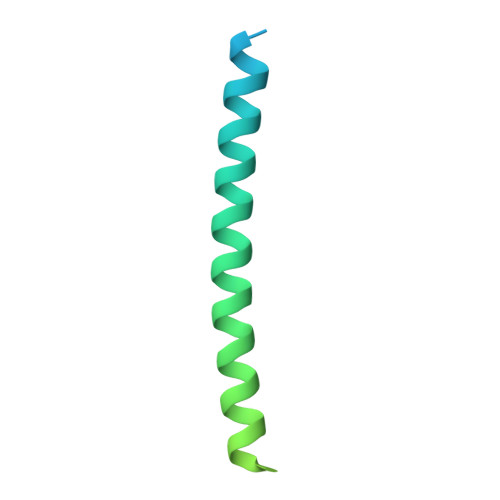 |
UniProt | |||||
Find proteins for P20411 (Rattus norvegicus) Explore P20411 Go to UniProtKB: P20411 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P20411 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| Y01 (Subject of Investigation/LOI) Query on Y01 | E [auth B] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |