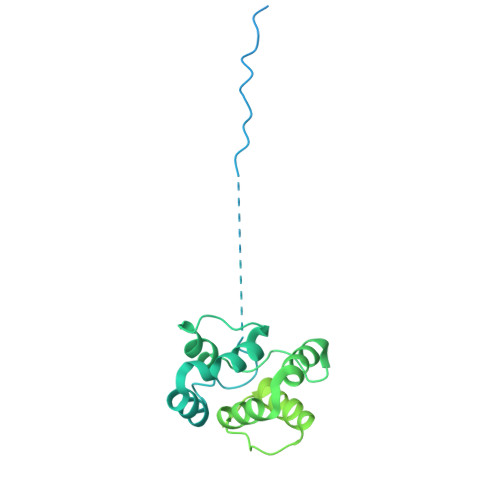
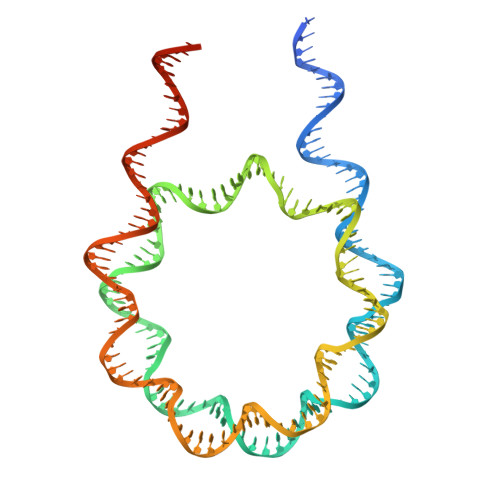
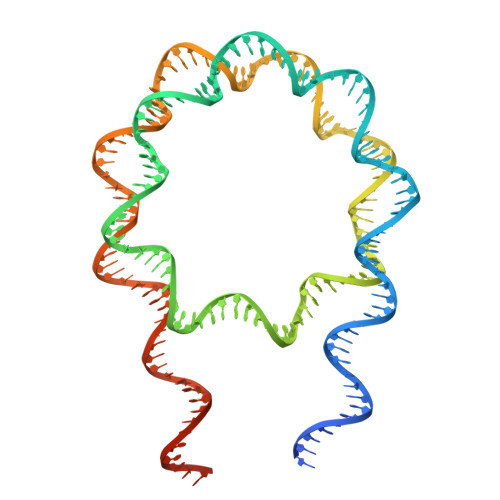
Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Kujirai, T., Echigoya, K., Kishi, Y., Saeki, M., Ito, T., Kato, J., Negishi, L., Kimura, H., Masumoto, H., Takizawa, Y., Gotoh, Y., Kurumizaka, H.(2025) Nat Struct Mol Biol 32: 1183-1192
- PubMed: 39984731
- DOI: https://doi.org/10.1038/s41594-025-01493-w
- Primary Citation of Related Structures:
8KCY, 8KD1, 8KE0 - PubMed Abstract:
Structural diversity of the nucleosome affects chromatin conformations and regulates eukaryotic genome functions. Here we identify DEK, whose function is unknown, as a nucleosome-binding protein. In embryonic neural progenitor cells, DEK colocalizes with H3 K27 trimethylation (H3K27me3), the facultative heterochromatin mark. DEK stimulates the methyltransferase activity of Polycomb repressive complex 2 (PRC2), which is responsible for H3K27me3 deposition in vitro. Cryo-electron microscopy structures of the DEK-nucleosome complexes reveal that DEK binds the nucleosome by its tripartite DNA-binding mode on the dyad and linker DNAs and interacts with the nucleosomal acidic patch by its newly identified histone-binding region. The DEK-nucleosome interaction mediates linker DNA reorientation and induces chromatin compaction, which may facilitate PRC2 activation. These findings provide mechanistic insights into chromatin structure-mediated gene regulation by DEK.
- Laboratory of Chromatin Structure and Function, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: