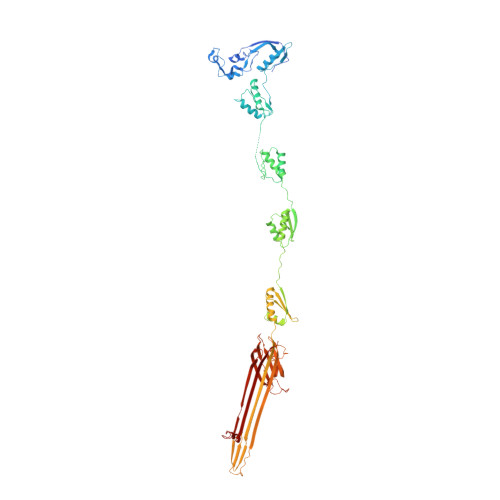
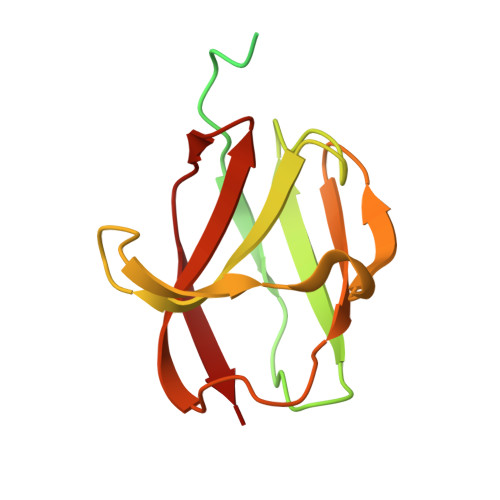
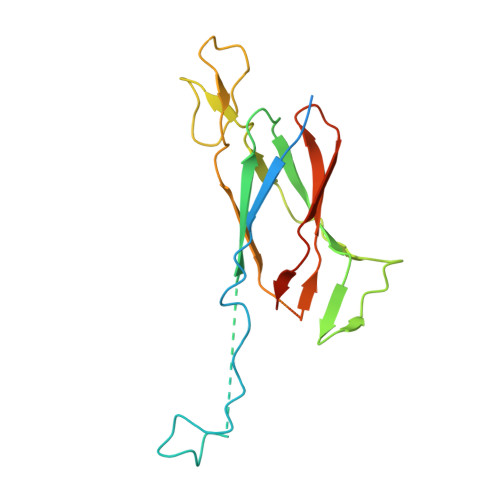
Structural characterization and functional insights into the type II secretion system of the poly-extremophile Deinococcus radiodurans.
Farci, D., Milenkovic, S., Iesu, L., Tanas, M., Ceccarelli, M., Piano, D.(2024) J Biological Chem 300: 105537-105537
- PubMed: 38072042
- DOI: https://doi.org/10.1016/j.jbc.2023.105537
- Primary Citation of Related Structures:
8CO1 - PubMed Abstract:
The extremophile bacterium D. radiodurans boasts a distinctive cell envelope characterized by the regular arrangement of three protein complexes. Among these, the Type II Secretion System (T2SS) stands out as a pivotal structural component. We used cryo-electron microscopy to reveal unique features, such as an unconventional protein belt (DR_1364) around the main secretin (GspD), and a cap (DR_0940) found to be a separated subunit rather than integrated with GspD. Furthermore, a novel region at the N-terminus of the GspD constitutes an additional second gate, supplementing the one typically found in the outer membrane region. This T2SS was found to contribute to envelope integrity, while also playing a role in nucleic acid and nutrient trafficking. Studies on intact cell envelopes show a consistent T2SS structure repetition, highlighting its significance within the cellular framework.
- Department of Plant Physiology, Warsaw University of Life Sciences - SGGW, Warsaw, Poland; Department of Life and Environmental Sciences, Università degli Studi di Cagliari, Cagliari, Italy; R&D Department, ReGenFix Laboratories, Sardara, Italy. Electronic address: domenica.farci@unica.it.
Organizational Affiliation: