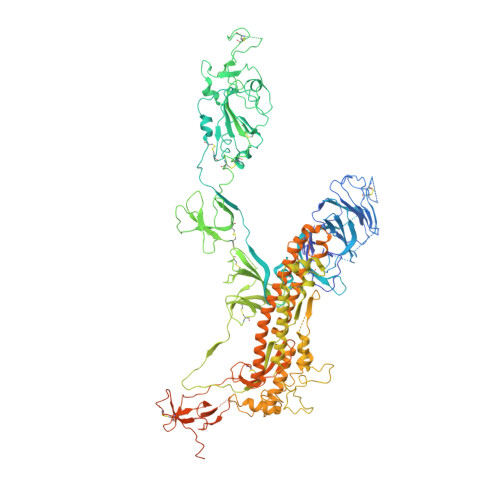
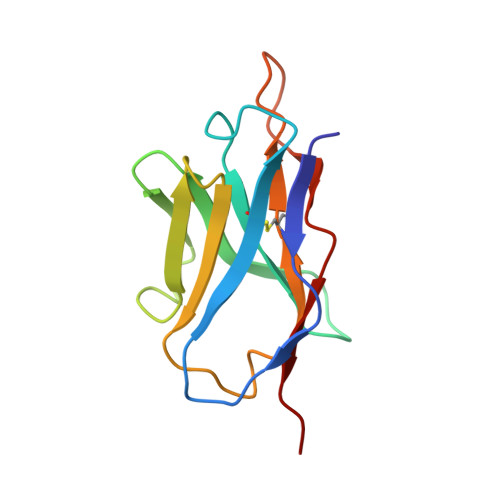
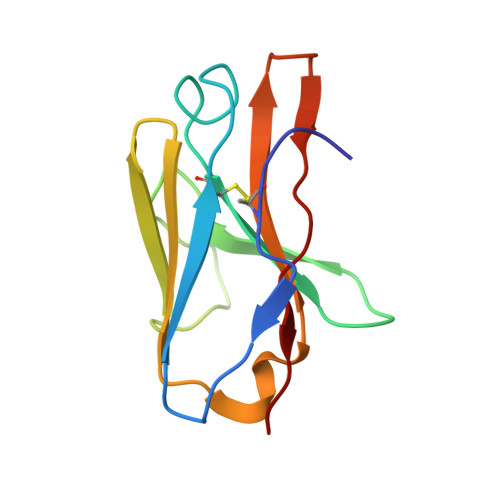
The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Chen, X., Mohapatra, A., Nguyen, H.T.V., Schimanski, L., Kit Tan, T., Rijal, P., Chen, C.P., Cheng, S.H., Lee, W.H., Chou, Y.C., Townsend, A.R., Ma, C., Huang, K.A.(2024) PLoS Pathog 20: e1012246-e1012246
- PubMed: 38857264
- DOI: https://doi.org/10.1371/journal.ppat.1012246
- Primary Citation of Related Structures:
8X0X, 8X0Y, 8YRO, 8YRP, 8YZ5, 8YZ6 - PubMed Abstract:
Antibody-mediated immunity plays a key role in protection against SARS-CoV-2. We characterized B-cell-derived anti-SARS-CoV-2 RBD antibody repertoires from vaccinated and infected individuals and elucidate the mechanism of action of broadly neutralizing antibodies and dissect antibodies at the epitope level. The breadth and clonality of anti-RBD B cell response varies among individuals. The majority of neutralizing antibody clones lose or exhibit reduced activities against Beta, Delta, and Omicron variants. Nevertheless, a portion of anti-RBD antibody clones that develops after a primary series or booster dose of COVID-19 vaccination exhibit broad neutralization against emerging Omicron BA.2, BA.4, BA.5, BQ.1.1, XBB.1.5 and XBB.1.16 variants. These broadly neutralizing antibodies share genetic features including a conserved usage of the IGHV3-53 and 3-9 genes and recognize three clustered epitopes of the RBD, including epitopes that partially overlap the classically defined set identified early in the pandemic. The Fab-RBD crystal and Fab-Spike complex structures corroborate the epitope grouping of antibodies and reveal the detailed binding mode of broadly neutralizing antibodies. Structure-guided mutagenesis improves binding and neutralization potency of antibody with Omicron variants via a single amino-substitution. Together, these results provide an immunological basis for partial protection against severe COVID-19 by the ancestral strain-based vaccine and indicate guidance for next generation monoclonal antibody development and vaccine design.
Organizational Affiliation:
Genomics Research Center, Academia Sinica, Taipei, Taiwan.