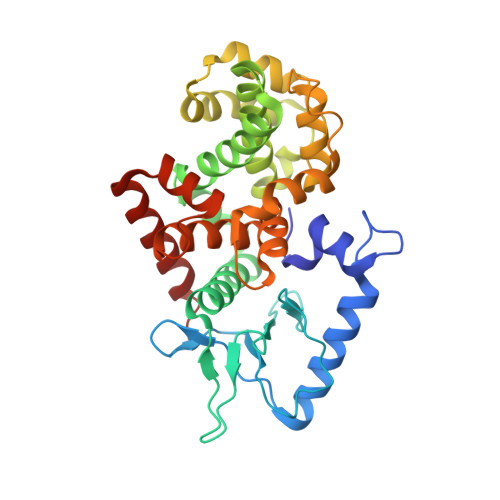
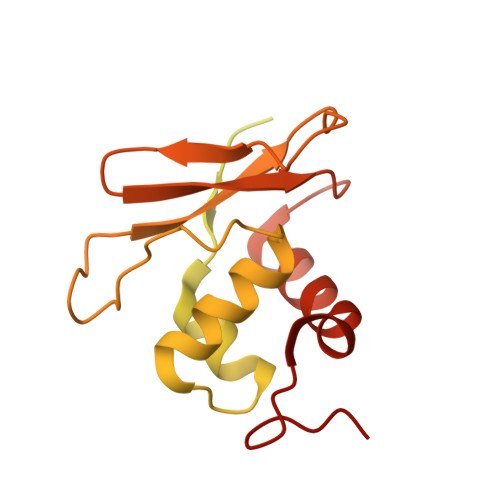
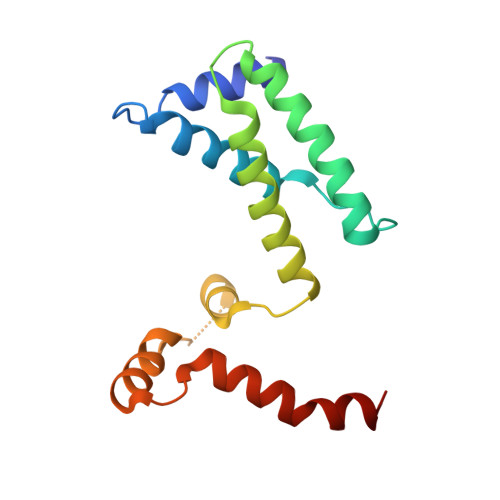
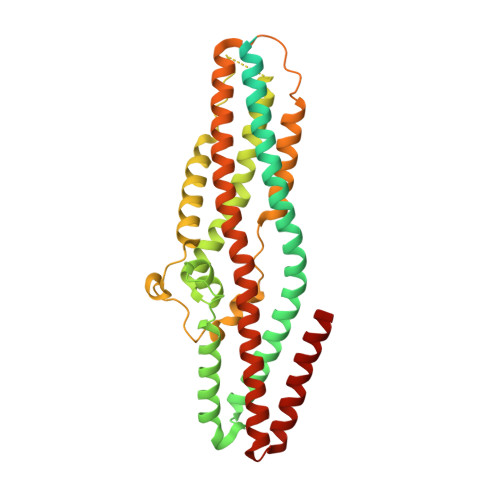
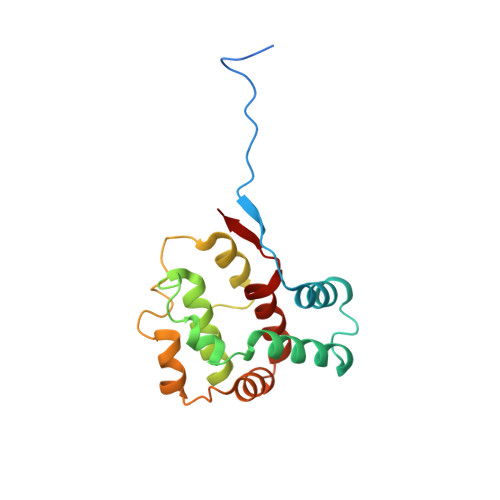
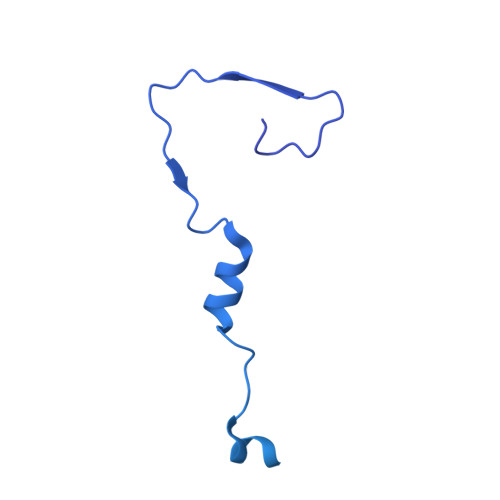
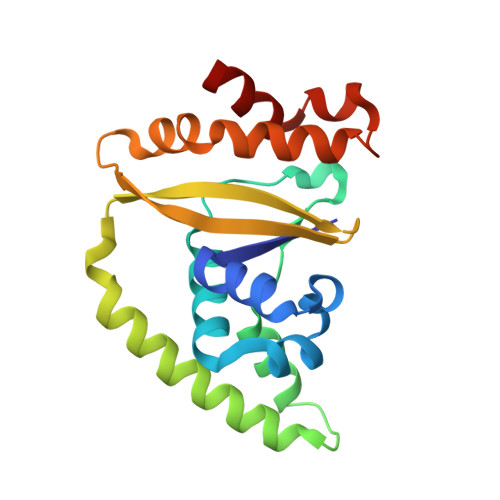
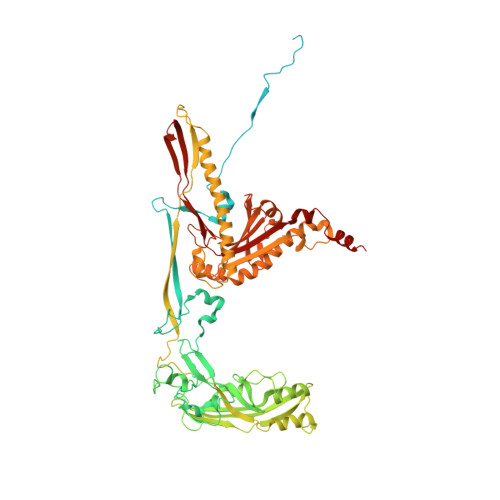
Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Yang, Y., Shao, Q., Guo, M., Han, L., Zhao, X., Wang, A., Li, X., Wang, B., Pan, J.A., Chen, Z., Fokine, A., Sun, L., Fang, Q.(2024) Nat Commun 15: 6551-6551
- PubMed: 39095371
- DOI: https://doi.org/10.1038/s41467-024-50811-1
- Primary Citation of Related Structures:
8Y6V - PubMed Abstract:
Jumbo phages are a group of tailed bacteriophages with large genomes and capsids. As a prototype of jumbo phage, ΦKZ infects Pseudomonas aeruginosa, a multi-drug-resistant (MDR) opportunistic pathogen leading to acute or chronic infection in immunocompromised individuals. It holds potential to be used as an antimicrobial agent and as a model for uncovering basic phage biology. Although previous low-resolution structural studies have indicated that jumbo phages may have more complicated capsid structures than smaller phages such as HK97, the detailed structures and the assembly mechanism of their capsids remain largely unknown. Here, we report a 3.5-Å-resolution cryo-EM structure of the ΦKZ capsid. The structure unveiled ten minor capsid proteins, with some decorating the outer surface of the capsid and the others forming a complex network attached to the capsid's inner surface. This network seems to play roles in driving capsid assembly and capsid stabilization. Similar mechanisms of capsid assembly and stabilization are probably employed by many other jumbo viruses.
Organizational Affiliation:
School of Public Health (Shenzhen), Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong, China.