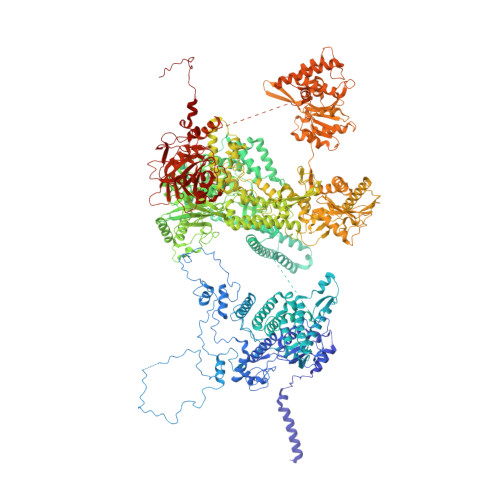
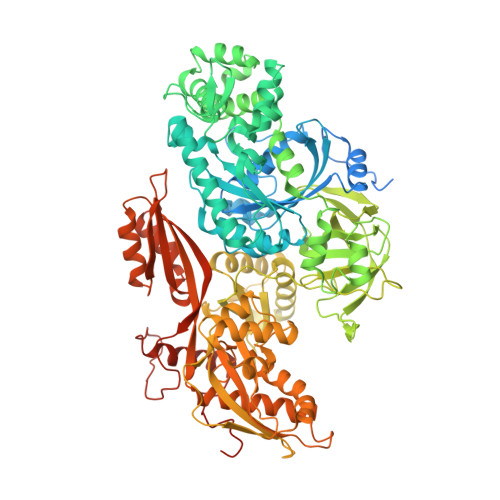
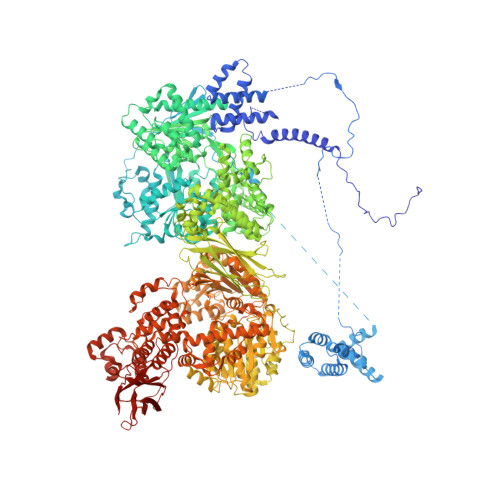
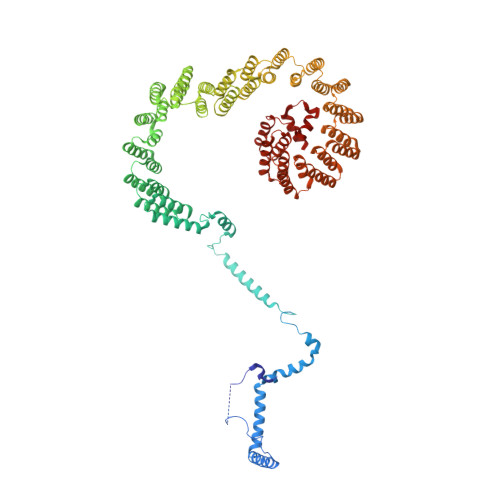
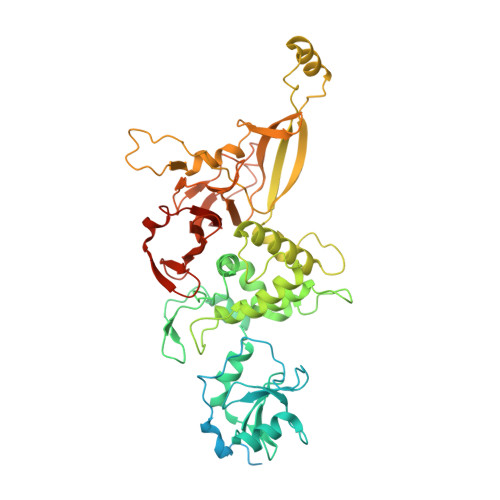
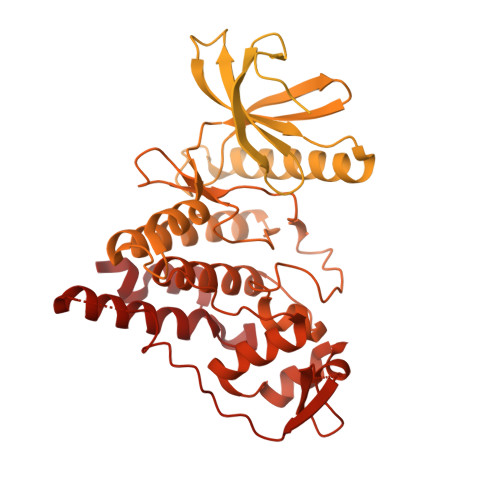
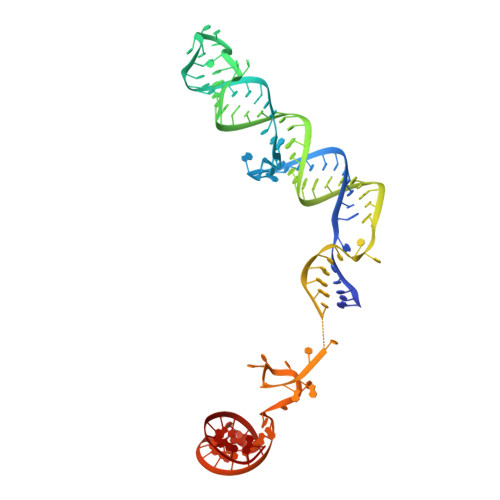
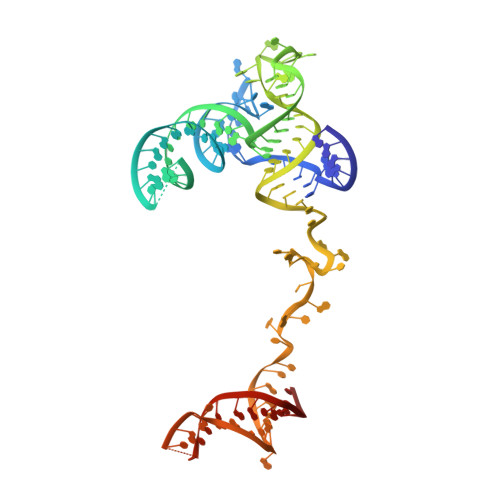
Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Bai, R., Yuan, M., Zhang, P., Luo, T., Shi, Y., Wan, R.(2024) Science 383: 1245-1252
- PubMed: 38484052
- DOI: https://doi.org/10.1126/science.adn7272
- Primary Citation of Related Structures:
8Y6O, 8Y7E - PubMed Abstract:
The minor spliceosome, which is responsible for the splicing of U12-type introns, comprises five small nuclear RNAs (snRNAs), of which only one is shared with the major spliceosome. In this work, we report the 3.3-angstrom cryo-electron microscopy structure of the fully assembled human minor spliceosome pre-B complex. The atomic model includes U11 small nuclear ribonucleoprotein (snRNP), U12 snRNP, and U4atac/U6atac.U5 tri-snRNP. U11 snRNA is recognized by five U11-specific proteins (20K, 25K, 35K, 48K, and 59K) and the heptameric Sm ring. The 3' half of the 5'-splice site forms a duplex with U11 snRNA; the 5' half is recognized by U11-35K, U11-48K, and U11 snRNA. Two proteins, CENATAC and DIM2/TXNL4B, specifically associate with the minor tri-snRNP. A structural analysis uncovered how two conformationally similar tri-snRNPs are differentiated by the minor and major prespliceosomes for assembly.
Organizational Affiliation:
Research Center for Industries of the Future, Key Zhejiang Key Laboratory of Structural Biology, School of Life Sciences, Westlake University, Xihu District, Hangzhou 310024, Zhejiang Province, China.