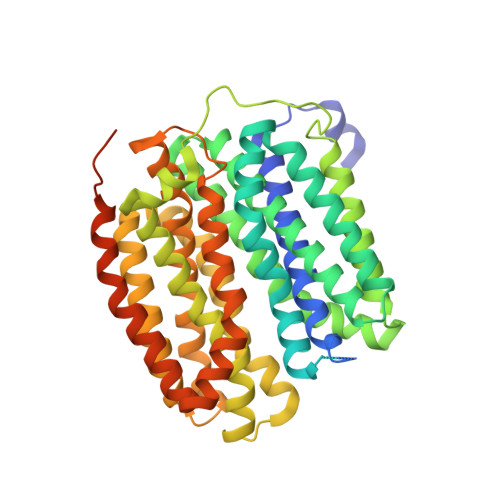
Neurotransmitter recognition by human vesicular monoamine transporter 2.
Im, D., Jormakka, M., Juge, N., Kishikawa, J.I., Kato, T., Sugita, Y., Noda, T., Uemura, T., Shiimura, Y., Miyaji, T., Asada, H., Iwata, S.(2024) Nat Commun 15: 7661-7661
- PubMed: 39284862
- DOI: https://doi.org/10.1038/s41467-024-51960-z
- Primary Citation of Related Structures:
8WRD, 8WRE, 8WVG - PubMed Abstract:
Human vesicular monoamine transporter 2 (VMAT2), a member of the SLC18 family, plays a crucial role in regulating neurotransmitters in the brain by facilitating their uptake and storage within vesicles, preparing them for exocytotic release. Because of its central role in neurotransmitter signalling and neuroprotection, VMAT2 is a target for neurodegenerative diseases and movement disorders, with its inhibitor being used as therapeutics. Despite the importance of VMAT2 in pharmacophysiology, the molecular basis of VMAT2-mediated neurotransmitter transport and its inhibition remains unclear. Here we show the cryo-electron microscopy structure of VMAT2 in the substrate-free state, in complex with the neurotransmitter dopamine, and in complex with the inhibitor tetrabenazine. In addition to these structural determinations, monoamine uptake assays, mutational studies, and pKa value predictions were performed to characterize the dynamic changes in VMAT2 structure. These results provide a structural basis for understanding VMAT2-mediated vesicular transport of neurotransmitters and a platform for modulation of current inhibitor design.
Organizational Affiliation:
Department of Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto, Japan. im.dohyun.3s@kyoto-u.ac.jp.