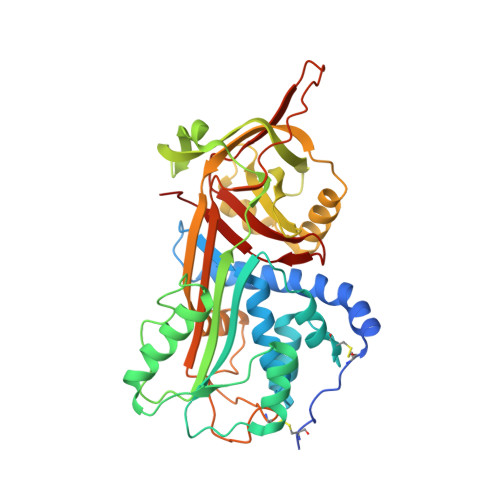
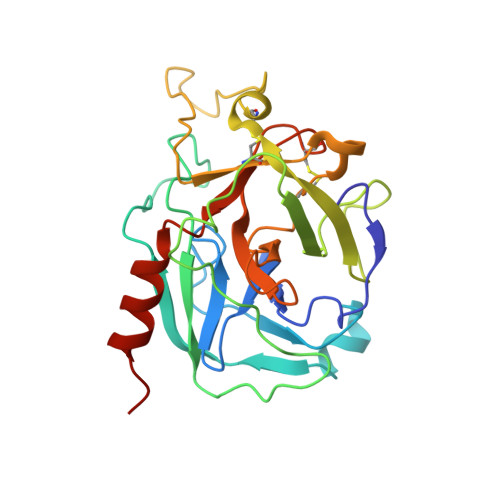
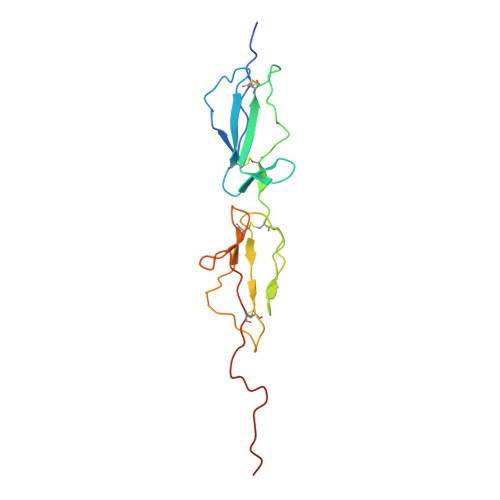
The Crystal Structure of the Michaelis-Menten Complex of C1 Esterase Inhibitor and C1s Reveals Novel Insights into Complement Regulation.
Garrigues, R.J., Garrison, M.P., Garcia, B.L.(2024) J Immunol
- PubMed: 38995166
- DOI: https://doi.org/10.4049/jimmunol.2400194
- Primary Citation of Related Structures:
8W18 - PubMed Abstract:
The ancient arm of innate immunity known as the complement system is a blood proteolytic cascade involving dozens of membrane-bound and solution-phase components. Although many of these components serve as regulatory molecules to facilitate controlled activation of the cascade, C1 esterase inhibitor (C1-INH) is the sole canonical complement regulator belonging to a superfamily of covalent inhibitors known as serine protease inhibitors (SERPINs). In addition to its namesake role in complement regulation, C1-INH also regulates proteases of the coagulation, fibrinolysis, and contact pathways. Despite this, the structural basis for C1-INH recognition of its target proteases has remained elusive. In this study, we present the crystal structure of the Michaelis-Menten (M-M) complex of the catalytic domain of complement component C1s and the SERPIN domain of C1-INH at a limiting resolution of 3.94 Å. Analysis of the structure revealed that nearly half of the protein/protein interface is formed by residues outside of the C1-INH reactive center loop. The contribution of these residues to the affinity of the M-M complex was validated by site-directed mutagenesis using surface plasmon resonance. Parallel analysis confirmed that C1-INH-interfacing residues on C1s surface loops distal from the active site also drive affinity of the M-M complex. Detailed structural comparisons revealed differences in substrate recognition by C1s compared with C1-INH recognition and highlight the importance of exosite interactions across broader SERPIN/protease systems. Collectively, this study improves our understanding of how C1-INH regulates the classical pathway of complement, and it sheds new light on how SERPINs recognize their cognate protease targets.
Organizational Affiliation:
Department of Microbiology and Immunology, Brody School of Medicine, East Carolina University, Greenville, NC.