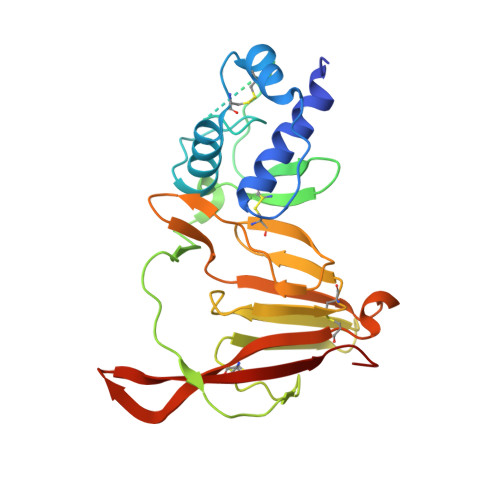
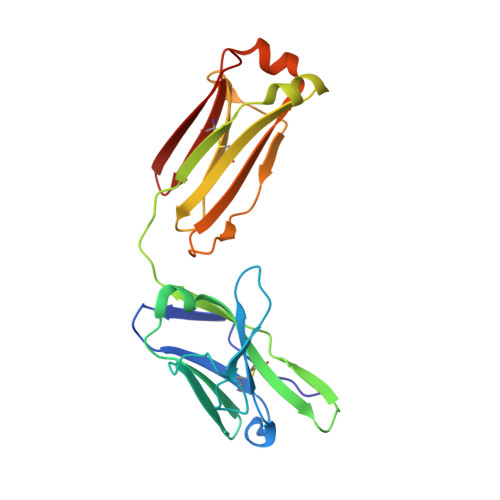
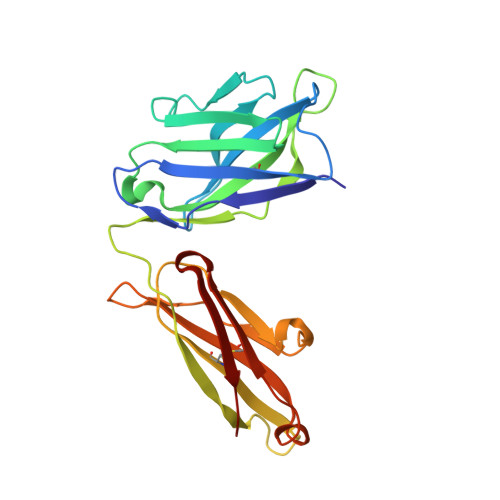
Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Shin, O.S., Monticelli, S.R., Hjorth, C.K., Hornet, V., Doyle, M., Abelson, D., Kuehne, A.I., Wang, A., Bakken, R.R., Mishra, A.K., Middlecamp, M., Champney, E., Stuart, L., Maurer, D.P., Li, J., Berrigan, J., Barajas, J., Balinandi, S., Lutwama, J.J., Lobel, L., Zeitlin, L., Walker, L.M., Dye, J.M., Chandran, K., Herbert, A.S., Pauli, N.T., McLellan, J.S.(2024) Cell Rep 43: 114502-114502
- PubMed: 39002130
- DOI: https://doi.org/10.1016/j.celrep.2024.114502
- Primary Citation of Related Structures:
8VVK, 8VVL, 8VWW - PubMed Abstract:
Crimean-Congo hemorrhagic fever virus can cause lethal disease in humans yet there are no approved medical countermeasures. Viral glycoprotein GP38, exclusive to Nairoviridae, is a target of protective antibodies and is a key antigen in preclinical vaccine candidates. Here, we isolate 188 GP38-specific antibodies from human survivors of infection. Competition experiments show that these antibodies bind across 5 distinct antigenic sites, encompassing 11 overlapping regions. Additionally, we show structures of GP38 bound with 9 of these antibodies targeting different antigenic sites. Although these GP38-specific antibodies are non-neutralizing, several display protective efficacy equal to or better than murine antibody 13G8 in two highly stringent rodent models of infection. Together, these data expand our understanding regarding this important viral protein and may inform the development of broadly effective CCHFV antibody therapeutics.
Organizational Affiliation:
Adimab, LLC, Lebanon, NH 03766, USA.