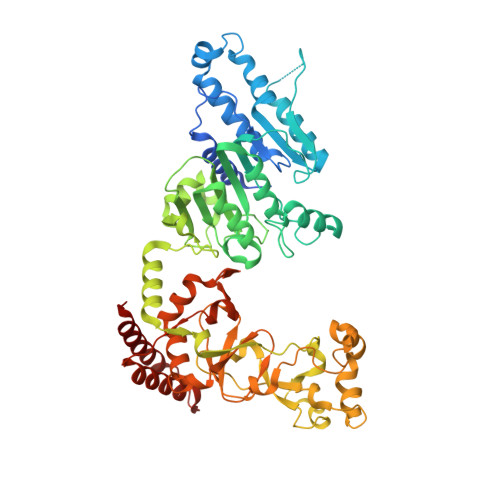
Quinoline-based compounds can inhibit diverse enzymes that act on DNA.
Zhou, J., Chen, Q., Ren, R., Yang, J., Liu, B., Horton, J.R., Chang, C., Li, C., Maksoud, L., Yang, Y., Rotili, D., Zhang, X., Blumenthal, R.M., Chen, T., Gao, Y., Valente, S., Mai, A., Cheng, X.(2024) bioRxiv
- PubMed: 38617249
- DOI: https://doi.org/10.1101/2024.04.03.587980
- Primary Citation of Related Structures:
8VPG, 8VPH, 8VPI - PubMed Abstract:
DNA methylation, as exemplified by cytosine-C5 methylation in mammals and adenine-N6 methylation in bacteria, is a crucial epigenetic mechanism driving numerous vital biological processes. Developing non-nucleoside inhibitors to cause DNA hypomethylation is a high priority, in order to treat a variety of significant medical conditions without the toxicities associated with existing cytidine-based hypomethylating agents. In this study, we have characterized fifteen quinoline-based analogs. Notably, compounds with additions like a methylamine ( 9 ) or methylpiperazine ( 11 ) demonstrate similar low micromolar inhibitory potency against both human DNMT1 (which generates C5-methylcytosine) and Clostridioides difficile CamA (which generates N6-methyladenine). Structurally, compounds 9 and 11 specifically intercalate into CamA-bound DNA via the minor groove, adjacent to the target adenine, leading to a substantial conformational shift that moves the catalytic domain away from the DNA. This study adds to the limited examples of DNA methyltransferases being inhibited by non-nucleotide compounds through DNA intercalation, following the discovery of dicyanopyridine-based inhibitors for DNMT1. Furthermore, our study shows that some of these quinoline-based analogs inhibit other enzymes that act on DNA, such as polymerases and base excision repair glycosylases. Finally, in cancer cells compound 11 elicits DNA damage response via p53 activation. Six of fifteen quinoline-based derivatives demonstrated comparable low micromolar inhibitory effects on human cytosine methyltransferase DNMT1, and the bacterial adenine methyltransferases Clostridioides difficile CamA and Caulobacter crescentus CcrM. Compounds 9 and 11 were found to intercalate into a DNA substrate bound by CamA. These quinoline-based derivatives also showed inhibitory activity against various base excision repair DNA glycosylases, and DNA and RNA polymerases. Compound 11 provokes DNA damage response via p53 activation in cancer cells.