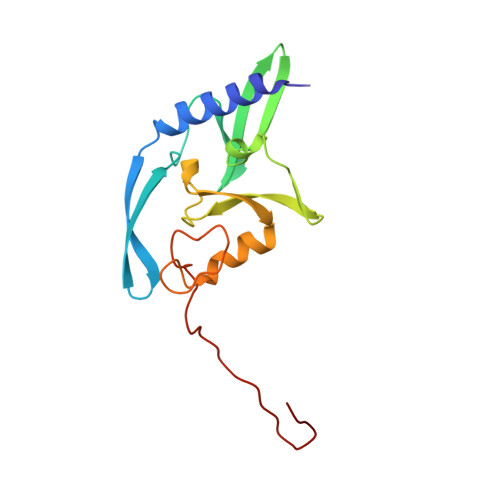
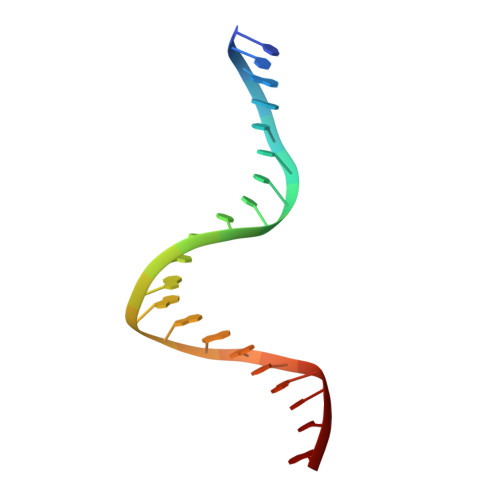
Observing one-divalent-metal-ion-dependent and histidine-promoted His-Me family I-PpoI nuclease catalysis in crystallo.
Chang, C., Zhou, G., Gao, Y.(2024) Elife 13
- PubMed: 39141555
- DOI: https://doi.org/10.7554/eLife.99960
- Primary Citation of Related Structures:
8VMO, 8VMP, 8VMQ, 8VMR, 8VMS, 8VMT, 8VMU, 8VMV, 8VMW, 8VMX, 8VMY, 8VMZ, 8VN0, 8VN1, 8VN2, 8VN3, 8VN4, 8VN5, 8VN6, 8VN7, 8VN8, 8VN9, 8VNA, 8VNB, 8VNC, 8VND, 8VNE, 8VNF, 8VNG, 8VNH, 8VNJ, 8VNK, 8VNL, 8VNM, 8VNN, 8VNO, 8VNP, 8VNQ, 8VNR, 8VNS, 8VNT, 8VNU - PubMed Abstract:
Metal-ion-dependent nucleases play crucial roles in cellular defense and biotechnological applications. Time-resolved crystallography has resolved catalytic details of metal-ion-dependent DNA hydrolysis and synthesis, uncovering the essential roles of multiple metal ions during catalysis. The histidine-metal (His-Me) superfamily nucleases are renowned for binding one divalent metal ion and requiring a conserved histidine to promote catalysis. Many His-Me family nucleases, including homing endonucleases and Cas9 nuclease, have been adapted for biotechnological and biomedical applications. However, it remains unclear how the single metal ion in His-Me nucleases, together with the histidine, promotes water deprotonation, nucleophilic attack, and phosphodiester bond breakage. By observing DNA hydrolysis in crystallo with His-Me I-PpoI nuclease as a model system, we proved that only one divalent metal ion is required during its catalysis. Moreover, we uncovered several possible deprotonation pathways for the nucleophilic water. Interestingly, binding of the single metal ion and water deprotonation are concerted during catalysis. Our results reveal catalytic details of His-Me nucleases, which is distinct from multi-metal-ion-dependent DNA polymerases and nucleases.
- Department of Biosciences, Rice University, Houston, United States.
Organizational Affiliation: