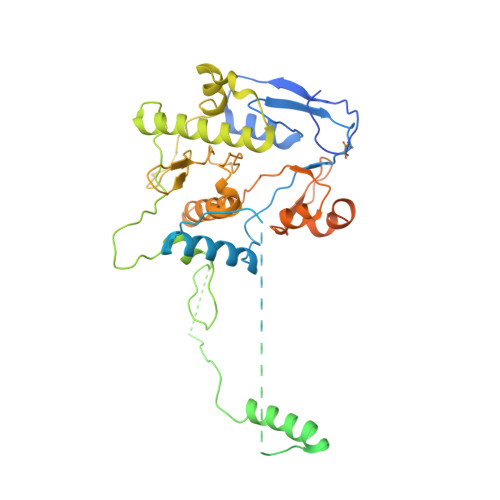
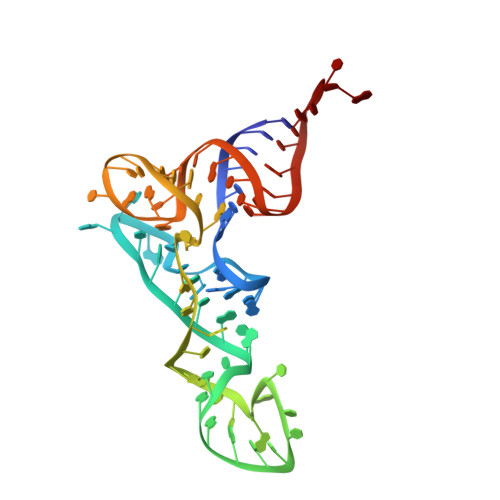
Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function.
Lan, X., Huang, W., Kim, S.B., Fu, D., Abeywansha, T., Lou, J., Balamurugan, U., Kwon, Y.T., Ji, C.H., Taylor, D.J., Zhang, Y.(2024) Nat Commun 15: 6350-6350
- PubMed: 39068213
- DOI: https://doi.org/10.1038/s41467-024-50719-w
- Primary Citation of Related Structures:
8TZV, 8UAU - PubMed Abstract:
The arginyl-transferase ATE1 is a tRNA-dependent enzyme that covalently attaches an arginine molecule to a protein substrate. Conserved from yeast to humans, ATE1 deficiency in mice correlates with defects in cardiovascular development and angiogenesis and results in embryonic lethality, while conditional knockouts exhibit reproductive, developmental, and neurological deficiencies. Despite the recent revelation of the tRNA binding mechanism and the catalytic cycle of yeast ATE1, the structure-function relationship of ATE1 in higher organisms is not well understood. In this study, we present the three-dimensional structure of human ATE1 in an apo-state and in complex with its tRNA cofactor and a peptide substrate. In contrast to its yeast counterpart, human ATE1 forms a symmetric homodimer, which dissociates upon binding of a substrate. Furthermore, human ATE1 includes a unique and extended loop that wraps around tRNA Arg , creating extensive contacts with the T-arm of the tRNA cofactor. Substituting key residues identified in the substrate binding site of ATE1 abolishes enzymatic activity and results in the accumulation of ATE1 substrates in cells.
Organizational Affiliation:
Department of Biochemistry, Case Western Reserve University, Cleveland, OH, USA.