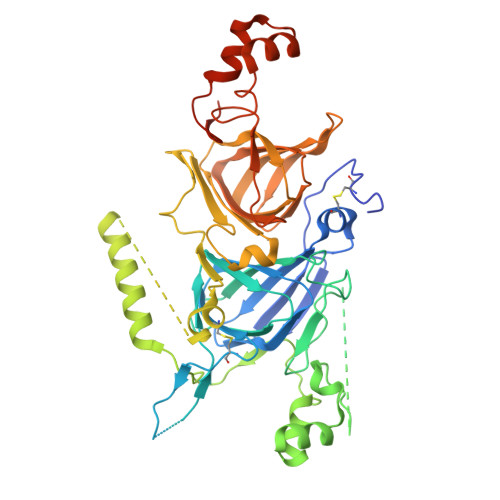
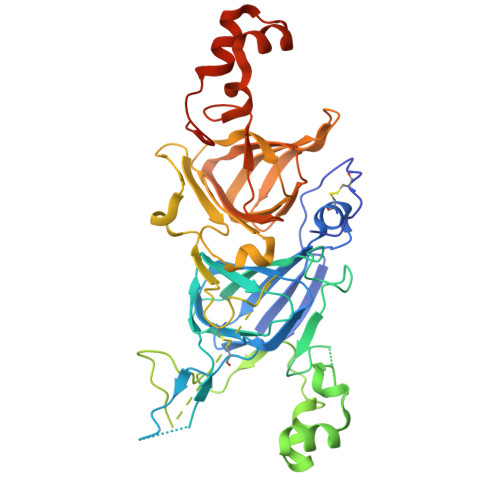
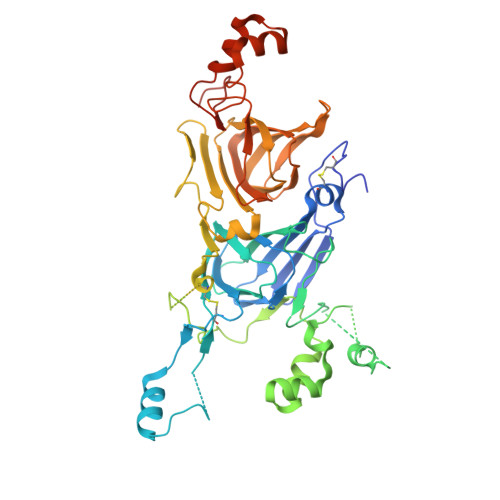
Crystal structure of hetero hexameric 11S seed storage protein of hazelnut.
Guo, F., Zhang, Y., Howard, A., Xu, Y.(2024) Plant Physiol Biochem 210: 108653-108653
- PubMed: 38670029
- DOI: https://doi.org/10.1016/j.plaphy.2024.108653
- Primary Citation of Related Structures:
8UAI - PubMed Abstract:
Edible plant seeds provide a relatively inexpensive source of protein and make up a large part of nutrients for humans. Plant seeds accumulate storage proteins during seed development. Seed storage proteins act as a reserve of nutrition for seed germination and seedling growth. However, seed storage proteins may be allergenic, and the prevalence of food allergy has increased rapidly in recent years. The 11S globulins account for a significant number of known major food allergens. They are of interest to the public and the agricultural industry because of food safety concerns and the need for crop enhancement. We sought to determine the crystal structure of Cor a 9, the 11 S storage protein of hazelnut and a food allergen. The structure was refined to 1.92 Å, and the R and R free for the refined structure are 17.6% and 22.5%, respectively. The structure of Cor a 9 showed a hetero hexamer of an 11S seed storage protein for the first time. The hexamer was two trimers associated back-to-back. Two long alpha helixes at the C-terminal end of the acidic domain of one of the Cor a 9 isoforms lay at the trimer-trimer interface's groove. These data provided much-needed information about the allergenicity of the 11S seed proteins. The information may also facilitate a better understanding of the folding and transportation of 11S seed storage proteins.
Organizational Affiliation:
Discover Biotherapeutics, Exelixis Inc. 1851 Harbor Bay Parkway, Alameda, CA 94502, USA.