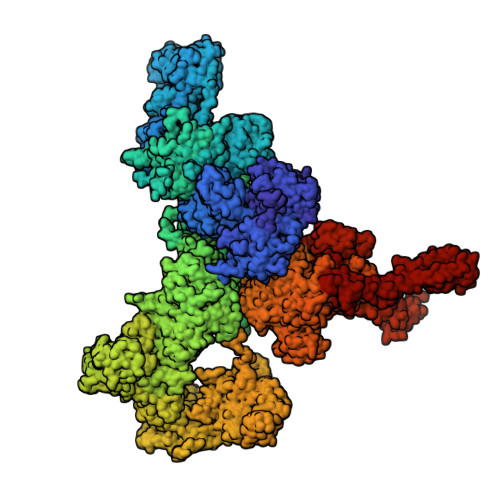
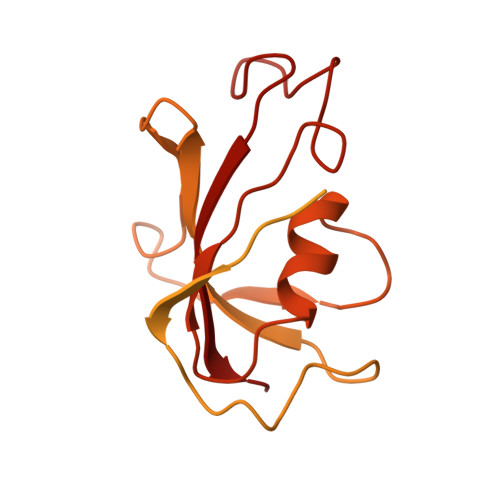
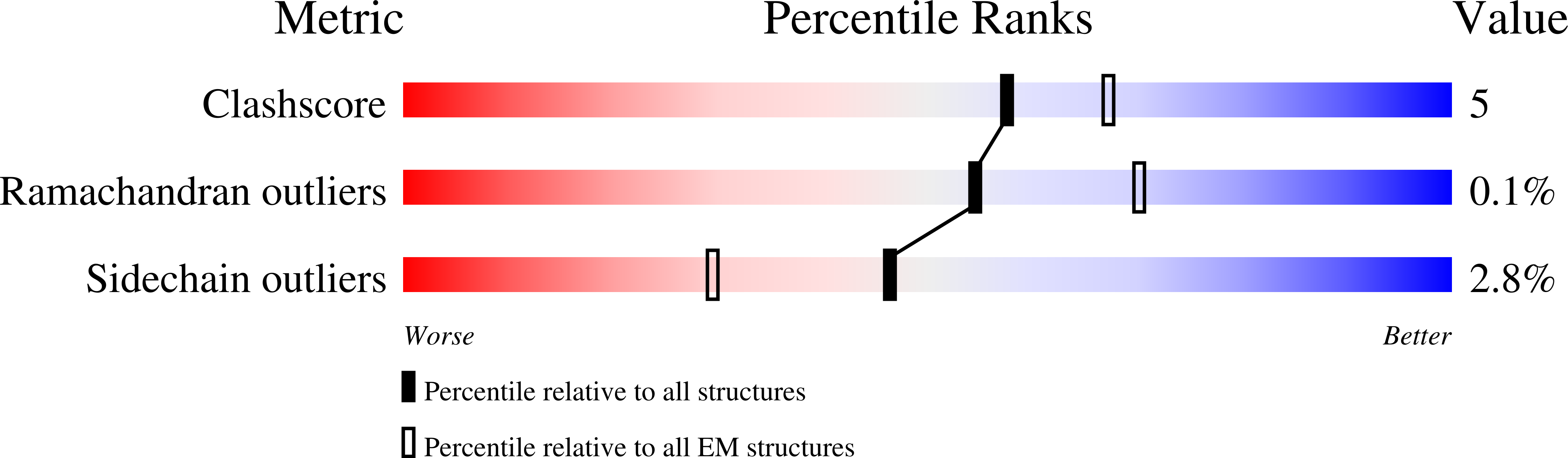Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Cholak, S., Saville, J.W., Zhu, X., Berezuk, A.M., Tuttle, K.S., Haji-Ghassemi, O., Alvarado, F.J., Van Petegem, F., Subramaniam, S.(2023) Structure 31: 790-800.e4
- PubMed: 37192614
- DOI: https://doi.org/10.1016/j.str.2023.04.009
- Primary Citation of Related Structures:
8SEN, 8SEO, 8SEP, 8SEQ, 8SER, 8SES, 8SET, 8SEU, 8SEV, 8SEW, 8SEX, 8SEY, 8SEZ, 8SF0 - PubMed Abstract:
The coordinated release of Ca 2+ from the sarcoplasmic reticulum (SR) is critical for excitation-contraction coupling. This release is facilitated by ryanodine receptors (RyRs) that are embedded in the SR membrane. In skeletal muscle, activity of RyR1 is regulated by metabolites such as ATP, which upon binding increase channel open probability (P o ). To obtain structural insights into the mechanism of RyR1 priming by ATP, we determined several cryo-EM structures of RyR1 bound individually to ATP-γ-S, ADP, AMP, adenosine, adenine, and cAMP. We demonstrate that adenine and adenosine bind RyR1, but AMP is the smallest ATP derivative capable of inducing long-range (>170 Å) structural rearrangements associated with channel activation, establishing a structural basis for key binding site interactions that are the threshold for triggering quaternary structural changes. Our finding that cAMP also induces these structural changes and results in increased channel opening suggests its potential role as an endogenous modulator of RyR1 conductance.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC V6T 1Z3, Canada.