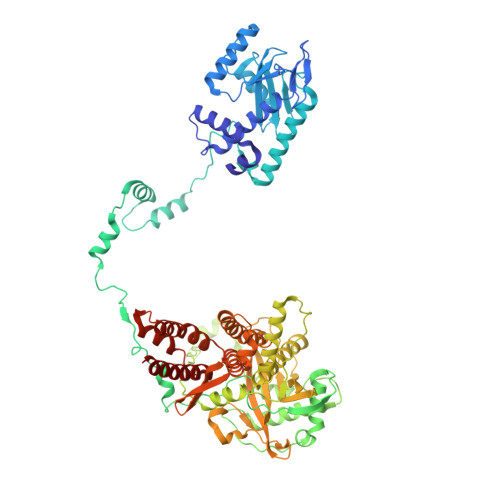
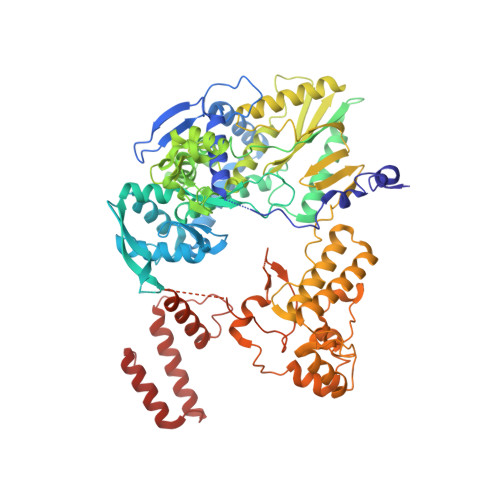
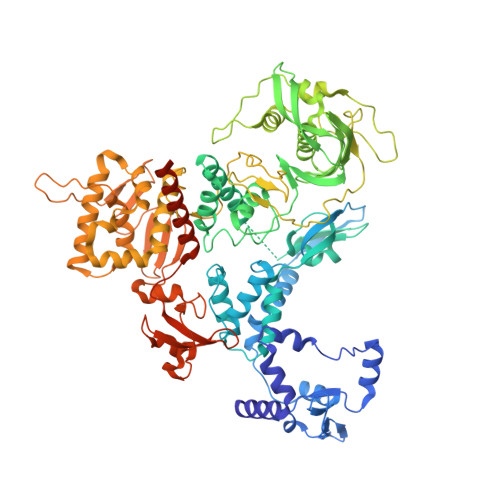
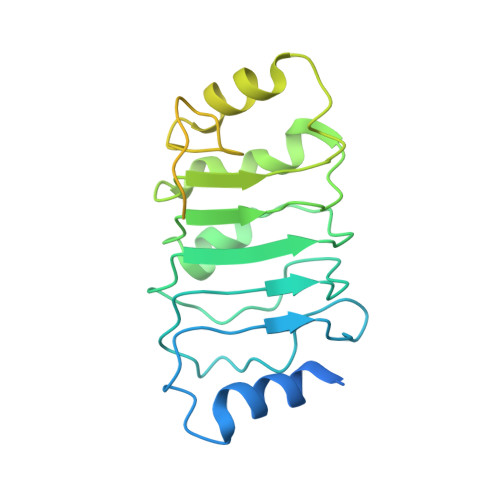
Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Arragain, B., Krischuns, T., Pelosse, M., Drncova, P., Blackledge, M., Naffakh, N., Cusack, S.(2024) Nat Commun 15: 6910-6910
- PubMed: 39160148
- DOI: https://doi.org/10.1038/s41467-024-51007-3
- Primary Citation of Related Structures:
8RMP, 8RMQ, 8RMR, 8RMS, 8RN0, 8RN1, 8RN2, 8RN3, 8RN4, 8RN5, 8RN6, 8RN7, 8RN8, 8RN9, 8RNA, 8RNB, 8RNC - PubMed Abstract:
Replication of influenza viral RNA depends on at least two viral polymerases, a parental replicase and an encapsidase, and cellular factor ANP32. ANP32 comprises an LRR domain and a long C-terminal low complexity acidic region (LCAR). Here we present evidence suggesting that ANP32 is recruited to the replication complex as an electrostatic chaperone that stabilises the encapsidase moiety within apo-polymerase symmetric dimers that are distinct for influenza A and B polymerases. The ANP32 bound encapsidase, then forms the asymmetric replication complex with the replicase, which is embedded in a parental ribonucleoprotein particle (RNP). Cryo-EM structures reveal the architecture of the influenza A and B replication complexes and the likely trajectory of the nascent RNA product into the encapsidase. The cryo-EM map of the FluB replication complex shows extra density attributable to the ANP32 LCAR wrapping around and stabilising the apo-encapsidase conformation. These structures give new insight into the various mutations that adapt avian strain polymerases to use the distinct ANP32 in mammalian cells.
Organizational Affiliation:
European Molecular Biology Laboratory, Grenoble, Cedex 9, France.