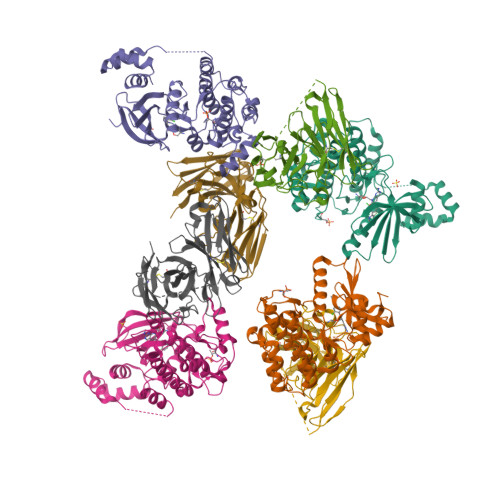
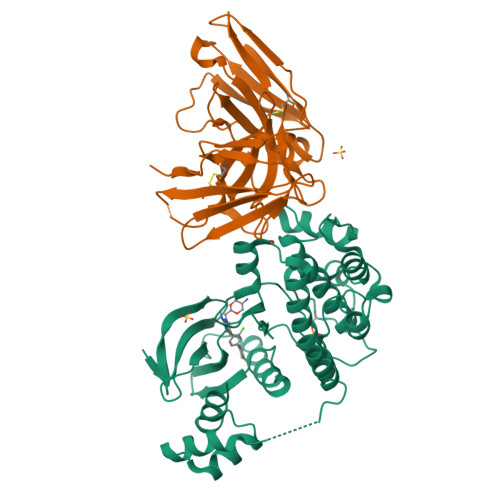
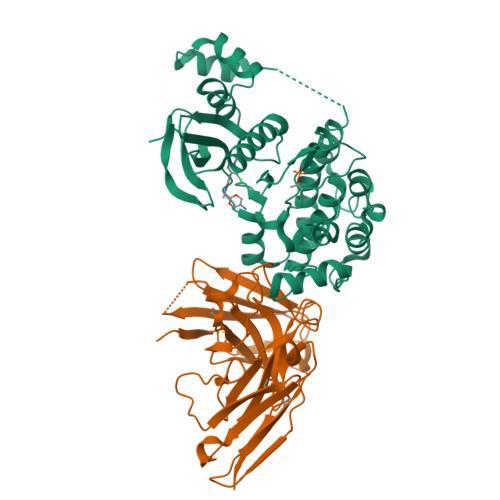
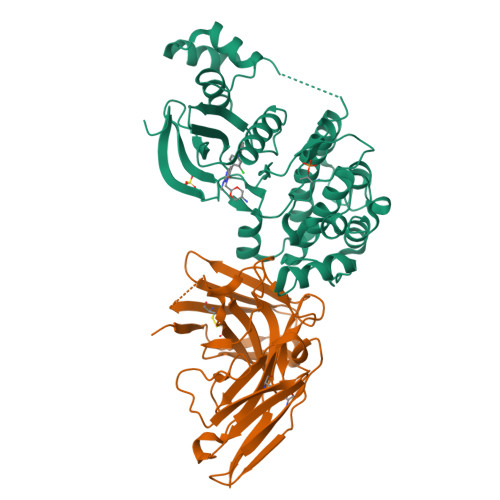
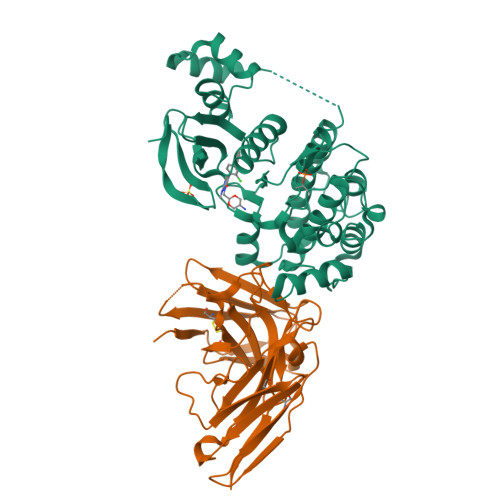
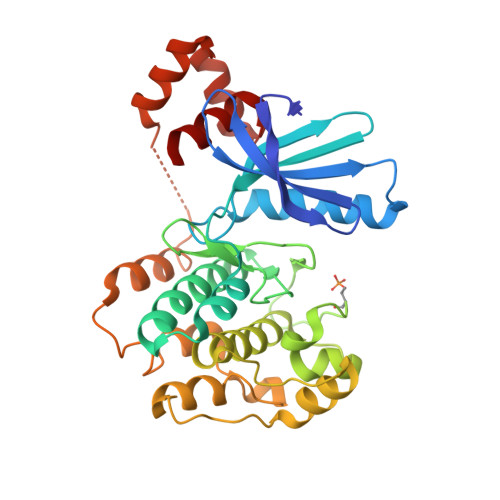
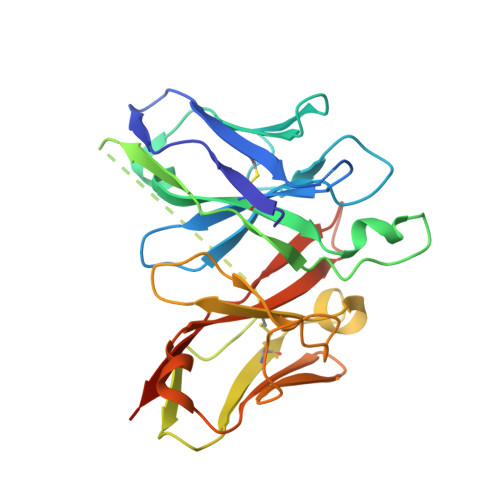
The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
Oster, L., Castaldo, M., de Vries, E., Edfeldt, F., Pemberton, N., Gordon, E., Cederblad, L., Kack, H.(2024) J Biological Chem 300: 107201-107201
- PubMed: 38508313
- DOI: https://doi.org/10.1016/j.jbc.2024.107201
- Primary Citation of Related Structures:
8R4O, 8R4Q, 8R4U, 8R4V - PubMed Abstract:
The salt-inducible kinases (SIKs) 1 to 3, belonging to the AMPK-related kinase family, serve as master regulators orchestrating a diverse set of physiological processes such as metabolism, bone formation, immune response, oncogenesis, and cardiac rhythm. Owing to its key regulatory role, the SIK kinases have emerged as compelling targets for pharmacological intervention across a diverse set of indications. Therefore, there is interest in developing SIK inhibitors with defined selectivity profiles both to further dissect the downstream biology and for treating disease. However, despite a large pharmaceutical interest in the SIKs, experimental structures of SIK kinases are scarce. This is likely due to the challenges associated with the generation of proteins suitable for structural studies. By adopting a rational approach to construct design and protein purification, we successfully crystallized and subsequently solved the structure of SIK3 in complex with HG-9-91-01, a potent SIK inhibitor. To enable further SIK3-inhibitor complex structures we identified an antibody fragment that facilitated crystallization and enabled a robust protocol suitable for structure-based drug design. The structures reveal SIK3 in an active conformation, where the ubiquitin-associated domain is shown to provide further stabilization to this active conformation. We present four pharmacologically relevant and distinct SIK3-inhibitor complexes. These detail the key interaction for each ligand and reveal how different regions of the ATP site are engaged by the different inhibitors to achieve high affinity. Notably, the structure of SIK3 in complex with a SIK3 specific inhibitor offers insights into isoform selectivity.
Organizational Affiliation:
Mechanistic and Structural Biology, Discovery Sciences, R&D, AstraZeneca, Gothenburg, Sweden. Electronic address: linda.oster@astrazeneca.com.