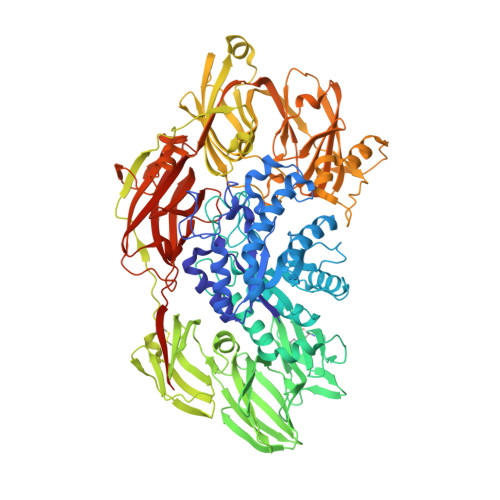
The archaeal highly thermostable GH35 family beta-galactosidase Da beta Gal has a unique seven domain protein fold.
Kil, Y., Pichkur, E.B., Sergeev, V.R., Zabrodskaya, Y., Myasnikov, A., Konevega, A.L., Shtam, T., Samygina, V.R., Rychkov, G.N.(2024) FEBS J 291: 3686-3705
- PubMed: 38825733
- DOI: https://doi.org/10.1111/febs.17166
- Primary Citation of Related Structures:
8QQH - PubMed Abstract:
The most extensively studied β-d-galactosidases (EC3.2.1.23) belonging to four glycoside hydrolase (GH) families 1, 2, 35, and 42 are widely distributed among Bacteria, Archaea and Eukaryotes. Here, we report a novel GH35 family β-galactosidase from the hyperthermophilic Thermoprotei archaeon Desulfurococcus amylolyticus (DaβGal). Unlike fungal monomeric six-domain β-galactosidases, the DaβGal enzyme is a dimer; it has an extra jelly roll domain D7 and three composite domains (D4, D5, and D6) that are formed by the distantly located polypeptide chain regions. The enzyme possesses a high specificity for β-d-galactopyranosides, and its distinguishing feature is the ability to cleave pNP-β-d-fucopyranoside. DaβGal efficiently catalyzes the hydrolysis of lactose at high temperatures, remains stable and active at 65 °С, and retains activity at 95 °С with a half-life time value equal to 73 min. These properties make archaeal DaβGal a more attractive candidate for biotechnology than the widely used fungal β-galactosidases.
Organizational Affiliation:
Department of Molecular and Radiation Biophysics, Petersburg Nuclear Physics Institute named by B.P.Konstantinov of National Research Center "Kurchatov Institute", Gatchina, Russia.