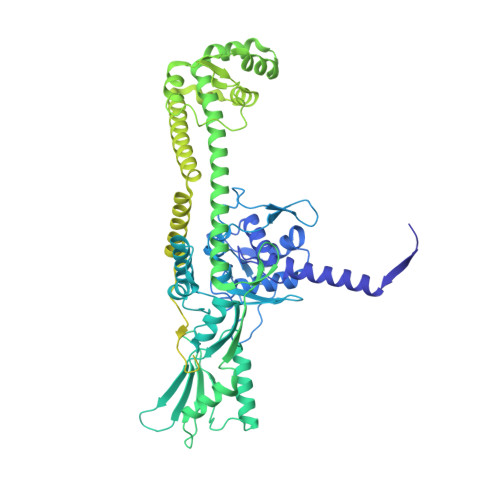
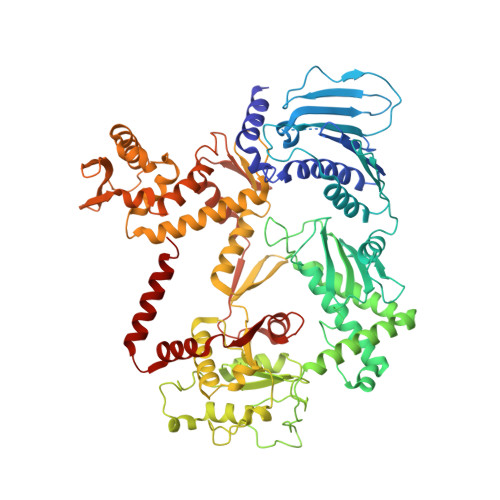
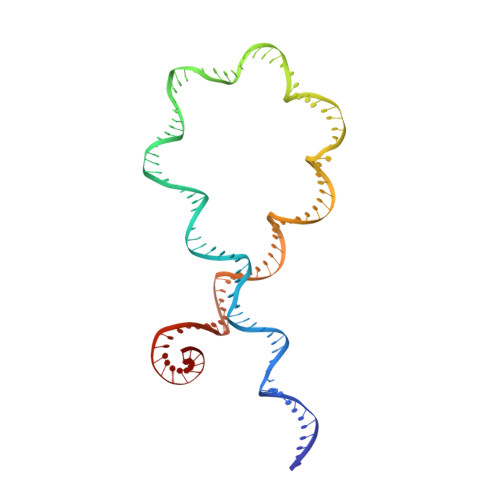
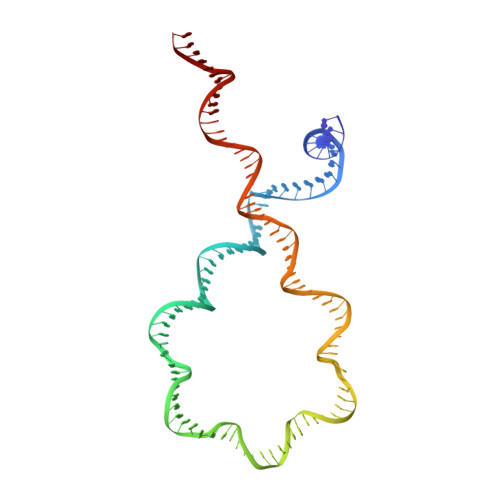
Structural basis of DNA crossover capture by Escherichia coli DNA gyrase.
Vayssieres, M., Marechal, N., Yun, L., Lopez Duran, B., Murugasamy, N.K., Fogg, J.M., Zechiedrich, L., Nadal, M., Lamour, V.(2024) Science 384: 227-232
- PubMed: 38603484
- DOI: https://doi.org/10.1126/science.adl5899
- Primary Citation of Related Structures:
8QDX, 8QQS, 8QQU - PubMed Abstract:
DNA supercoiling must be precisely regulated by topoisomerases to prevent DNA entanglement. The interaction of type IIA DNA topoisomerases with two DNA molecules, enabling the transport of one duplex through the transient double-stranded break of the other, remains elusive owing to structures derived solely from single linear duplex DNAs lacking topological constraints. Using cryo-electron microscopy, we solved the structure of Escherichia coli DNA gyrase bound to a negatively supercoiled minicircle DNA. We show how DNA gyrase captures a DNA crossover, revealing both conserved molecular grooves that accommodate the DNA helices. Together with molecular tweezer experiments, the structure shows that the DNA crossover is of positive chirality, reconciling the binding step of gyrase-mediated DNA relaxation and supercoiling in a single structure.
Organizational Affiliation:
Université de Strasbourg, Centre National de la Recherche Scientifique (CNRS), Institut national de la Recherche Médicale (INSERM), Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), UMR 7104- UMR-S 1258, F-67400 Illkirch, France.