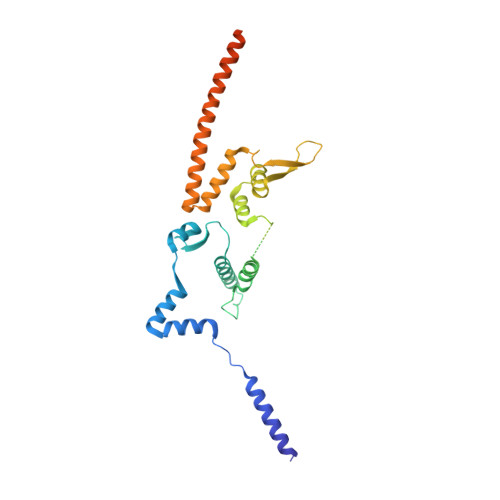
Structural mechanism of outer kinetochore Dam1-Ndc80 complex assembly on microtubules.
Muir, K.W., Batters, C., Dendooven, T., Yang, J., Zhang, Z., Burt, A., Barford, D.(2023) Science 382: 1184-1190
- PubMed: 38060647
- DOI: https://doi.org/10.1126/science.adj8736
- Primary Citation of Related Structures:
8Q84, 8Q85 - PubMed Abstract:
Kinetochores couple chromosomes to the mitotic spindle to segregate the genome during cell division. An error correction mechanism drives the turnover of kinetochore-microtubule attachments until biorientation is achieved. The structural basis for how kinetochore-mediated chromosome segregation is accomplished and regulated remains an outstanding question. In this work, we describe the cryo-electron microscopy structure of the budding yeast outer kinetochore Ndc80 and Dam1 ring complexes assembled onto microtubules. Complex assembly occurs through multiple interfaces, and a staple within Dam1 aids ring assembly. Perturbation of key interfaces suppresses yeast viability. Force-rupture assays indicated that this is a consequence of impaired kinetochore-microtubule attachment. The presence of error correction phosphorylation sites at Ndc80-Dam1 ring complex interfaces and the Dam1 staple explains how kinetochore-microtubule attachments are destabilized and reset.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK.