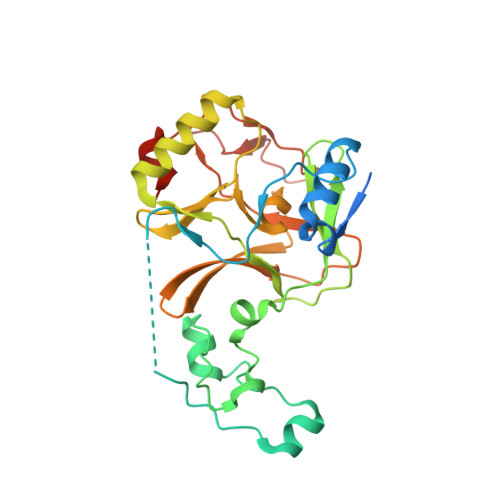
Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Le Coadou, L., Berthelet, J., Mechaly, A.E., Michail, C., Bui, L.C., Dairou, J., Haouz, A., Dupret, J.M., Rodrigues Lima, F.(2024) Biochem Biophys Res Commun 695: 149400-149400
- PubMed: 38160530
- DOI: https://doi.org/10.1016/j.bbrc.2023.149400
- Primary Citation of Related Structures:
8Q5P - PubMed Abstract:
SETD2 (SET-domain containing protein 2) is a histone methyltransferase (HMT) of the SET family responsible for the trimethylation of K36 of histone H3, thus producing the epigenetic mark H3K36me3. Recent studies have shown that certain SET family HMTs, such as SMYD2, SMYD3 or SETDB1 can also methylate protein kinases and therefore be involved in signaling pathways. Here we provide structural and enzymatic evidence showing that SETD2 methylates the protein tyrosine kinase ACK1 in vitro. ACK1 is recognized as a major integrator of signaling from various receptor tyrosine kinases. Using ACK1 peptides and recombinant proteins, we show that SETD2 methylates the K514 residue of ACK1 generating K514 mono, di or tri-methylation. Interestingly, K514 is found in a "H3K36-like" motif of ACK1 which is known to be post-translationally modified and to be involved in protein-protein interaction. The crystal structure of SETD2 catalytic domain in complex with an ACK1 peptide further provides the structural basis for the methylation of ACK1 K514 by SETD2. Our work therefore strongly suggests that ACK1 could be a novel non-histone substrate of SETD2 and further supports that SET HMTs, such as SETD2, could be involved in both epigenetic regulations and cell signaling.
Organizational Affiliation:
Université Paris Cité, CNRS, Unité de Biologie Fonctionnelle et Adaptative, F-75013, Paris, France.