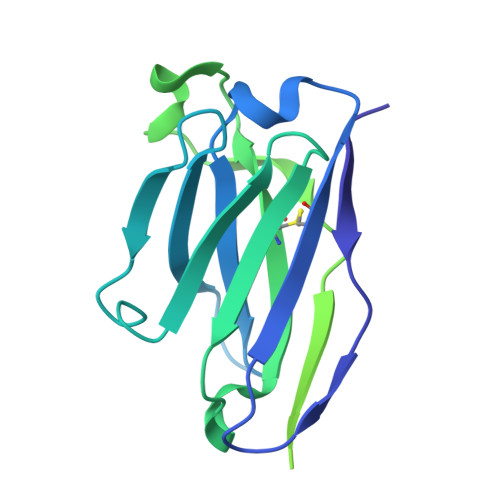
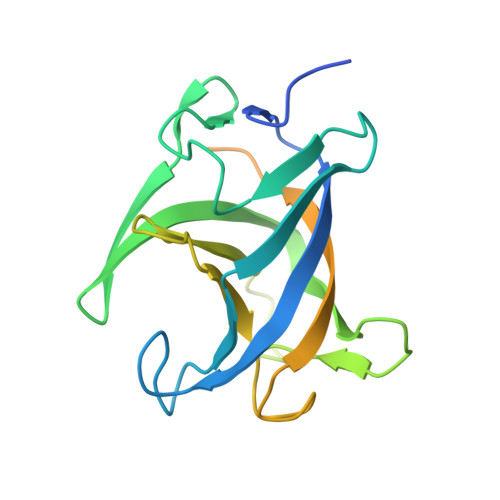
A novel class of broadly neutralizing hepatitis E virus-specific human antibodies
Ssebyatika, G., Dinkelborg, K., Stroeh, L.J., Hinte, F., Corneillie, L., Hueffner, L., Guzman, E.M., Nankya, P.L., Plueckebaum, N., Prallet, S., Mehnert, A.K., Verhoye, L., Juergens, C., Steinmann, E., Wedemeyer, H., Viejo-Borbolla, A., Dao Thi, L.V., Pietschmann, T., Luetgehetmann, M., Meuleman, P., Dandri, M., Krey, T., Behrendt, P.To be published.