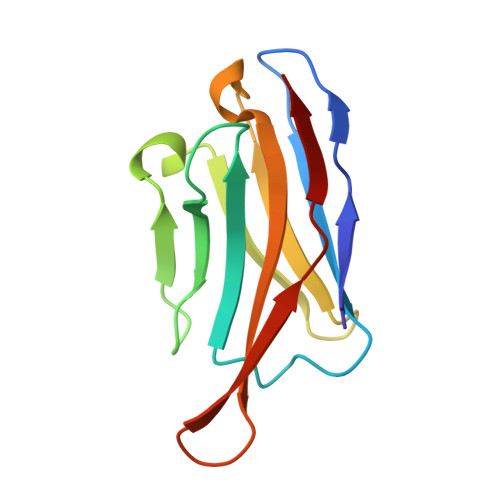
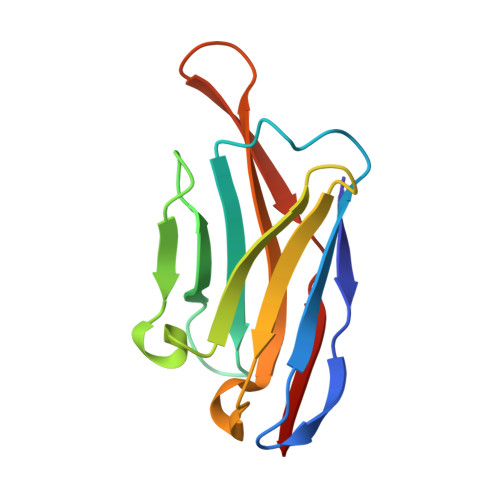
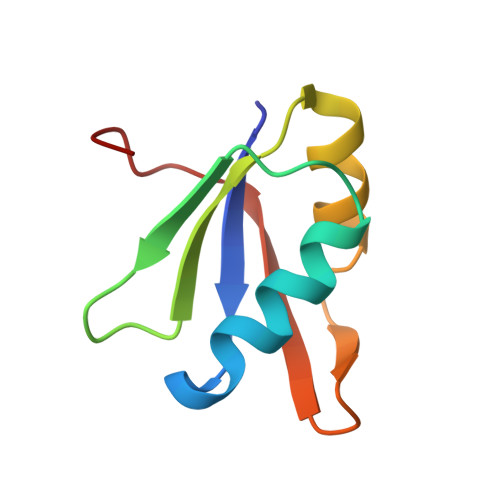
A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Sola Colom, M., Fu, Z., Gunkel, P., Guttler, T., Trakhanov, S., Srinivasan, V., Gregor, K., Pleiner, T., Gorlich, D.(2024) EMBO J 43: 2198-2232
- PubMed: 38649536
- DOI: https://doi.org/10.1038/s44318-024-00081-w
- Primary Citation of Related Structures:
7NOW, 7NQA, 7ZOX, 8CDS, 8CDT, 8OZB - PubMed Abstract:
Nuclear pore complex (NPC) biogenesis is a still enigmatic example of protein self-assembly. We now introduce several cross-reacting anti-Nup nanobodies for imaging intact nuclear pore complexes from frog to human. We also report a simplified assay that directly tracks postmitotic NPC assembly with added fluorophore-labeled anti-Nup nanobodies. During interphase, NPCs are inserted into a pre-existing nuclear envelope. Monitoring this process is challenging because newly assembled NPCs are indistinguishable from pre-existing ones. We overcame this problem by inserting Xenopus-derived NPCs into human nuclear envelopes and using frog-specific anti-Nup nanobodies for detection. We further asked whether anti-Nup nanobodies could serve as NPC assembly inhibitors. Using a selection strategy against conserved epitopes, we obtained anti-Nup93, Nup98, and Nup155 nanobodies that block Nup-Nup interfaces and arrest NPC assembly. We solved structures of nanobody-target complexes and identified roles for the Nup93 α-solenoid domain in recruiting Nup358 and the Nup214·88·62 complex, as well as for Nup155 and the Nup98 autoproteolytic domain in NPC scaffold assembly. The latter suggests a checkpoint linking pore formation to the assembly of the Nup98-dominated permeability barrier.
Organizational Affiliation:
Department of Cellular Logistics, Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany.