Structural basis of rotavirus spike proteolytic activation
Asensio-Cob, D., Perez-Mata, C., Gomez-Blanco, J., Vargas, J., Rodriguez, J.M., Luque, D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Inner capsid protein VP2 | 882 | Rotavirus | Mutation(s): 0 Gene Names: VP2 | 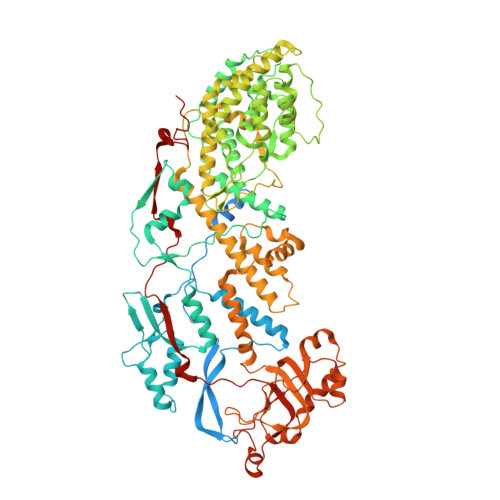 | |
UniProt | |||||
Find proteins for A2T3R1 (Rotavirus A) Explore A2T3R1 Go to UniProtKB: A2T3R1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A2T3R1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Outer capsid glycoprotein VP7 | 326 | Rotavirus | Mutation(s): 0 Gene Names: VP7, VP1 | 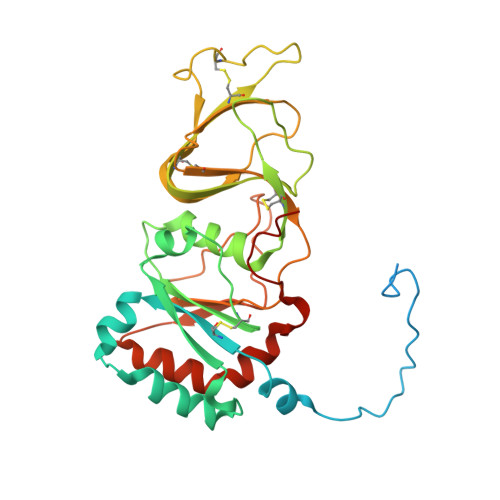 | |
UniProt | |||||
Find proteins for A0A060IEQ1 (Rotavirus A) Explore A0A060IEQ1 Go to UniProtKB: A0A060IEQ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A060IEQ1 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Intermediate capsid protein VP6 | 397 | Rotavirus | Mutation(s): 0 Gene Names: VP6 | 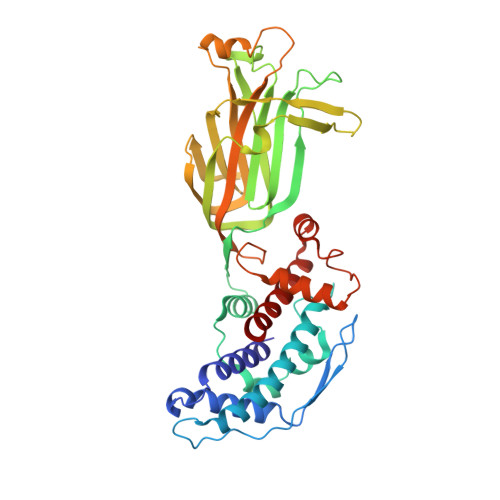 | |
UniProt | |||||
Find proteins for A2T3S6 (Rotavirus A) Explore A2T3S6 Go to UniProtKB: A2T3S6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A2T3S6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | AB [auth l] DB [auth m] EA [auth d] GB [auth n] HA [auth e] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| ZN Query on ZN | MB [auth C], NB [auth H], OB [auth I], PB [auth L], QB [auth O] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CA Query on CA | BB [auth l] CA [auth c] CB [auth l] DA [auth c] EB [auth m] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Spanish Ministry of Science, Innovation, and Universities | Spain | ISCIII-AESI PI20CIII/00014 |