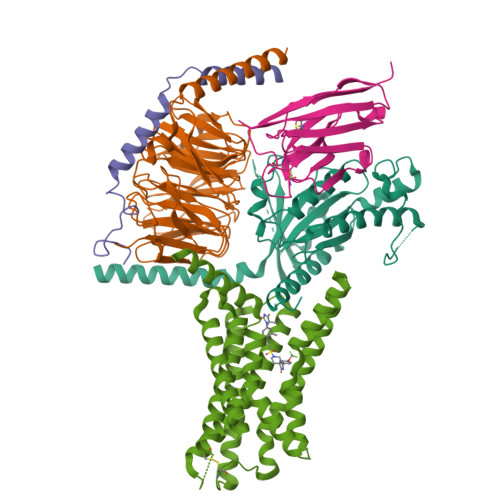
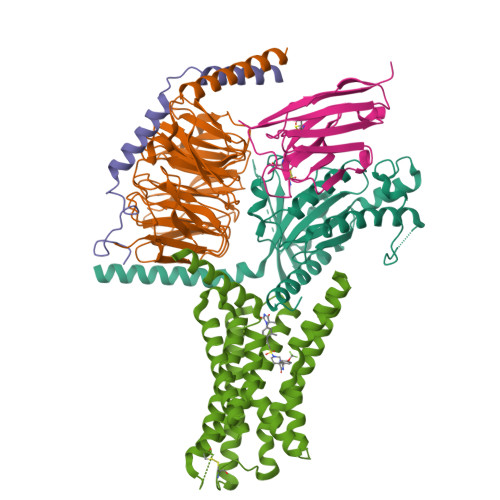
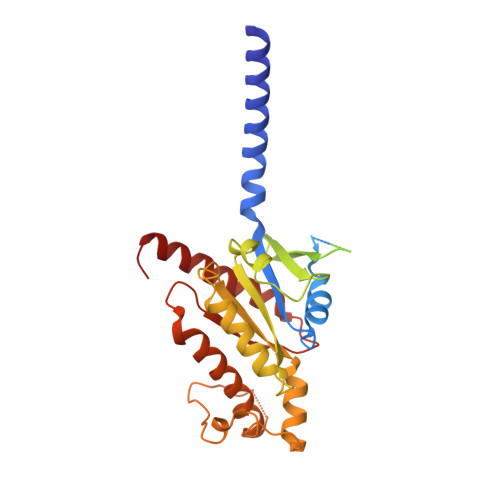
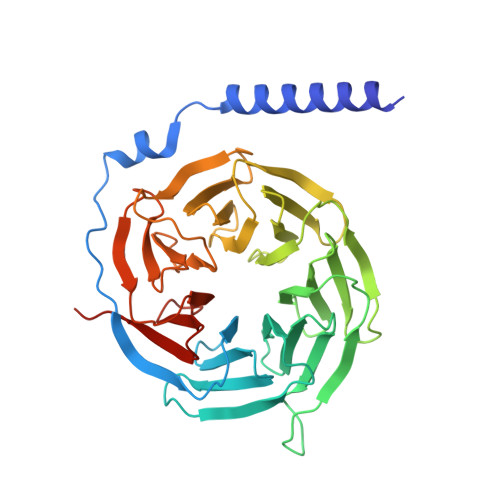
Conserved class B GPCR activation by a biased intracellular agonist.
Zhao, L.H., He, Q., Yuan, Q., Gu, Y., He, X., Shan, H., Li, J., Wang, K., Li, Y., Hu, W., Wu, K., Shen, J., Xu, H.E.(2023) Nature 621: 635-641
- PubMed: 37524305
- DOI: https://doi.org/10.1038/s41586-023-06467-w
- Primary Citation of Related Structures:
8JR9 - PubMed Abstract:
Class B G-protein-coupled receptors (GPCRs), including glucagon-like peptide 1 receptor (GLP1R) and parathyroid hormone 1 receptor (PTH1R), are important drug targets 1-5 . Injectable peptide drugs targeting these receptors have been developed, but orally available small-molecule drugs remain under development 6,7 . Here we report the high-resolution structure of human PTH1R in complex with the stimulatory G protein (G s ) and a small-molecule agonist, PCO371, which reveals an unexpected binding mode of PCO371 at the cytoplasmic interface of PTH1R with G s . The PCO371-binding site is totally different from all binding sites previously reported for small molecules or peptide ligands in GPCRs. The residues that make up the PCO371-binding pocket are conserved in class B GPCRs, and a single alteration in PTH2R and two residue alterations in GLP1R convert these receptors to respond to PCO371. Functional assays reveal that PCO371 is a G-protein-biased agonist that is defective in promoting PTH1R-mediated arrestin signalling. Together, these results uncover a distinct binding site for designing small-molecule agonists for PTH1R and possibly other members of the class B GPCRs and define a receptor conformation that is specific only for G-protein activation but not arrestin signalling. These insights should facilitate the design of distinct types of class B GPCR small-molecule agonist for various therapeutic indications.
Organizational Affiliation:
State Key Laboratory of Drug Research, Center for Structure and Function of Drug Targets, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China. zhaolihuawendy@simm.ac.cn.