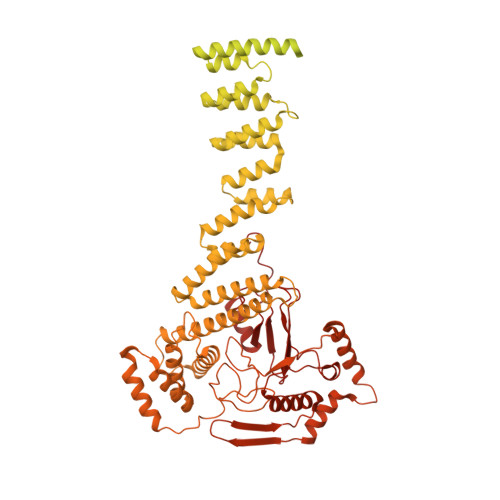
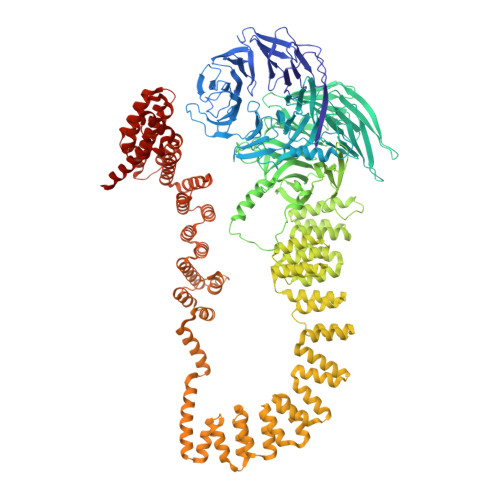
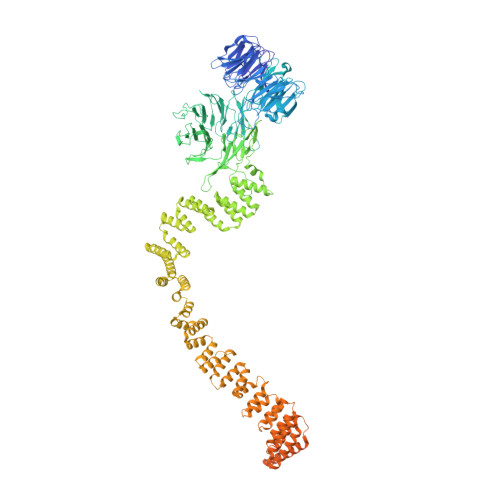
Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Ma, Y., He, J., Li, S., Yao, D., Huang, C., Wu, J., Lei, M.(2023) Nat Commun 14: 1506-1506
- PubMed: 36932088
- DOI: https://doi.org/10.1038/s41467-023-37208-2
- Primary Citation of Related Structures:
8HMC, 8HMD, 8HME, 8HMF - PubMed Abstract:
Intraflagellar transport (IFT) trains, the polymers composed of two multi-subunit complexes, IFT-A and IFT-B, carry out bidirectional intracellular transport in cilia, vital for cilia biogenesis and signaling. IFT-A plays crucial roles in the ciliary import of membrane proteins and the retrograde cargo trafficking. However, the molecular architecture of IFT-A and the assembly mechanism of the IFT-A into the IFT trains in vivo remains elusive. Here, we report the cryo-electron microscopic structures of the IFT-A complex from protozoa Tetrahymena thermophila. We find that IFT-A complexes present two distinct, elongated and folded states. Remarkably, comparison with the in situ cryo-electron tomography structure of the anterograde IFT train unveils a series of adjustments of the flexible arms in apo IFT-A when incorporated into the anterograde train. Our results provide an atomic-resolution model for the IFT-A complex and valuable insights into the assembly mechanism of anterograde IFT trains.
Organizational Affiliation:
Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200011, China.