The Type 9 Secretion System caught in the act of transport
Lauber, F., Deme, J.C., Liu, X., Kjaer, A., Miller, H.L., Alcock, F., Lea, S.M., Berks, B.C.(2024) Nat Microbiol
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) Nat Microbiol
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein involved in gliding motility SprA | 2,403 | Flavobacterium johnsoniae | Mutation(s): 0 | 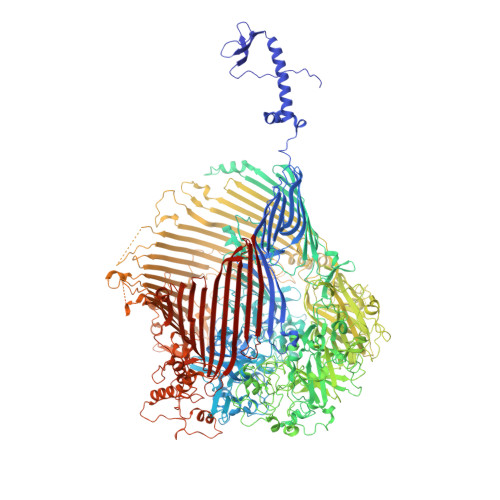 | |
UniProt | |||||
Find proteins for A0A1M5G5I4 (Flavobacterium johnsoniae) Explore A0A1M5G5I4 Go to UniProtKB: A0A1M5G5I4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1M5G5I4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptidyl-prolyl cis-trans isomerase | 176 | Flavobacterium johnsoniae | Mutation(s): 0 EC: 5.2.1.8 | 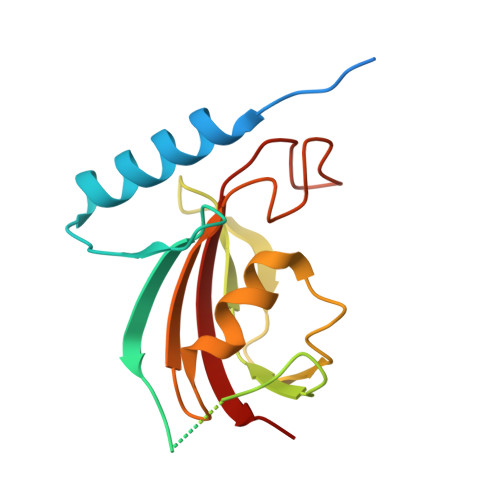 | |
UniProt | |||||
Find proteins for A5F9W9 (Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / BCRC 14874 / CCUG 350202 / NBRC 14942 / NCIMB 11054 / UW101)) Explore A5F9W9 Go to UniProtKB: A5F9W9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5F9W9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Type IX secretion system protein PorV domain-containing protein | C [auth F] | 402 | Flavobacterium johnsoniae | Mutation(s): 0 | 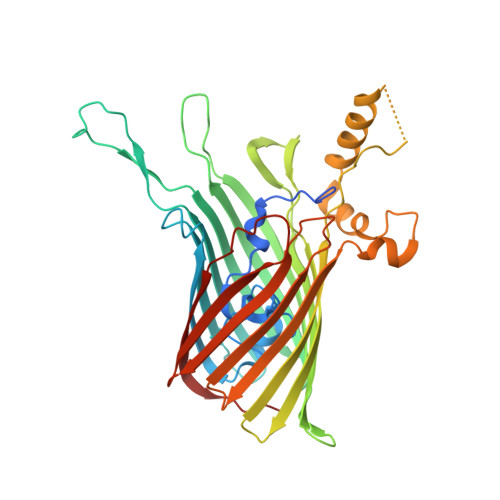 |
UniProt | |||||
Find proteins for A5FJM7 (Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / BCRC 14874 / CCUG 350202 / NBRC 14942 / NCIMB 11054 / UW101)) Explore A5FJM7 Go to UniProtKB: A5FJM7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5FJM7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RemZ | 1,114 | Flavobacterium johnsoniae | Mutation(s): 0 | 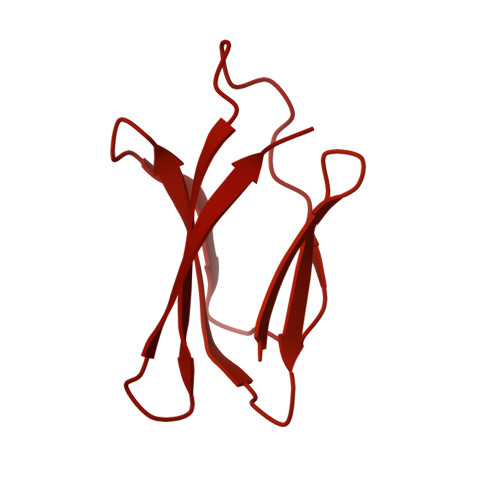 | |
UniProt | |||||
Find proteins for A5FLT3 (Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / BCRC 14874 / CCUG 350202 / NBRC 14942 / NCIMB 11054 / UW101)) Explore A5FLT3 Go to UniProtKB: A5FLT3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5FLT3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Periplasmic chaperone for outer membrane proteins Skp | E [auth G], F [auth C], G [auth E] | 341 | Flavobacterium johnsoniae | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1M5G3C1 (Flavobacterium johnsoniae) Explore A0A1M5G3C1 Go to UniProtKB: A0A1M5G3C1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1M5G3C1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SprE | H [auth I] | 870 | Flavobacterium johnsoniae | Mutation(s): 0 | 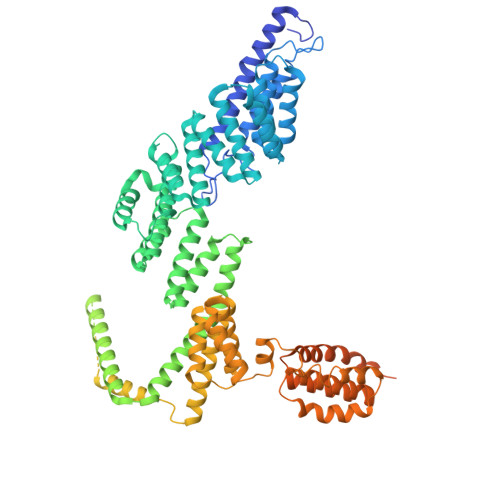 |
UniProt | |||||
Find proteins for A1E5T9 (Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / BCRC 14874 / CCUG 350202 / NBRC 14942 / NCIMB 11054 / UW101)) Explore A1E5T9 Go to UniProtKB: A1E5T9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A1E5T9 | ||||
Glycosylation | |||||
| Glycosylation Sites: 5 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LMN Query on LMN | I [auth A] | Lauryl Maltose Neopentyl Glycol C47 H88 O22 MADJBYLAYPCCOO-XYPZXBMFSA-N |  | ||
| MAN Query on MAN | J [auth I] K [auth I] L [auth I] M [auth I] N [auth I] | alpha-D-mannopyranose C6 H12 O6 WQZGKKKJIJFFOK-PQMKYFCFSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 3.0 |
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 219477/Z/19/Z |
| Wellcome Trust | United Kingdom | 107929/Z/15/Z |
| European Research Council (ERC) | European Union | 833713 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/S007474/1 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/S021264/1 |
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | CCR Core Funding for Lea Group |