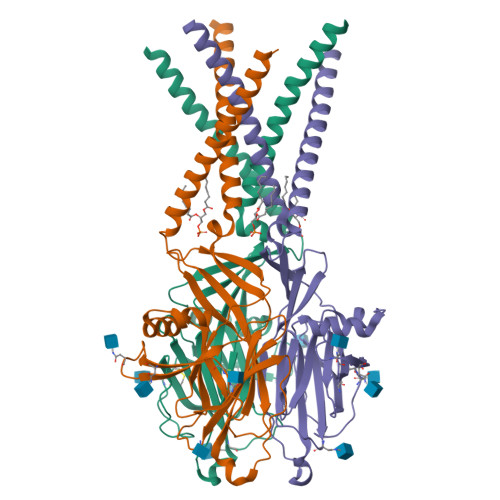
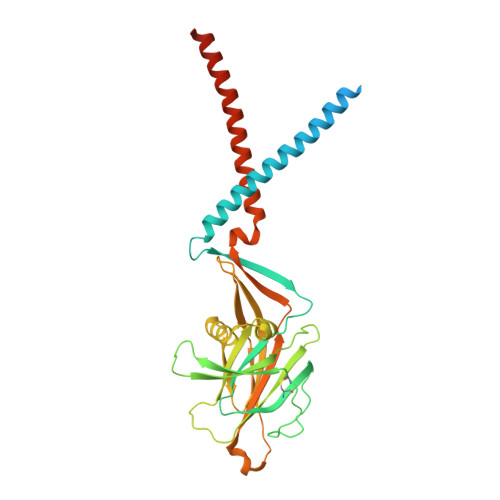
Inhibition of the proton-activated chloride channel PAC by PIP 2.
Mihaljevic, L., Ruan, Z., Osei-Owusu, J., Lu, W., Qiu, Z.(2023) Elife 12
- PubMed: 36633397
- DOI: https://doi.org/10.7554/eLife.83935
- Primary Citation of Related Structures:
8EQ4, 8FBL - PubMed Abstract:
Proton-activated chloride (PAC) channel is a ubiquitously expressed pH-sensing ion channel, encoded by PACC1 ( TMEM206 ). PAC regulates endosomal acidification and macropinosome shrinkage by releasing chloride from the organelle lumens. It is also found at the cell surface, where it is activated under pathological conditions related to acidosis and contributes to acid-induced cell death. However, the pharmacology of the PAC channel is poorly understood. Here, we report that phosphatidylinositol (4,5)-bisphosphate (PIP 2 ) potently inhibits PAC channel activity. We solved the cryo-electron microscopy structure of PAC with PIP 2 at pH 4.0 and identified its putative binding site, which, surprisingly, locates on the extracellular side of the transmembrane domain (TMD). While the overall conformation resembles the previously resolved PAC structure in the desensitized state, the TMD undergoes remodeling upon PIP 2 -binding. Structural and electrophysiological analyses suggest that PIP 2 inhibits the PAC channel by stabilizing the channel in a desensitized-like conformation. Our findings identify PIP 2 as a new pharmacological tool for the PAC channel and lay the foundation for future drug discovery targeting this channel.
Organizational Affiliation:
Department of Physiology, Johns Hopkins University School of Medicine, Baltimore, United States.