Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
Bothe, U., Gunther, J., Nubbemeyer, R., Siebeneicher, H., Ring, S., Bomer, U., Peters, M., Rausch, A., Denner, K., Himmel, H., Sutter, A., Terebesi, I., Lange, M., Wengner, A.M., Guimond, N., Thaler, T., Platzek, J., Eberspacher, U., Schafer, M., Steuber, H., Zollner, T.M., Steinmeyer, A., Schmidt, N.(2024) J Med Chem 67: 1225-1242
- PubMed: 38228402
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01714
- Primary Citation of Related Structures:
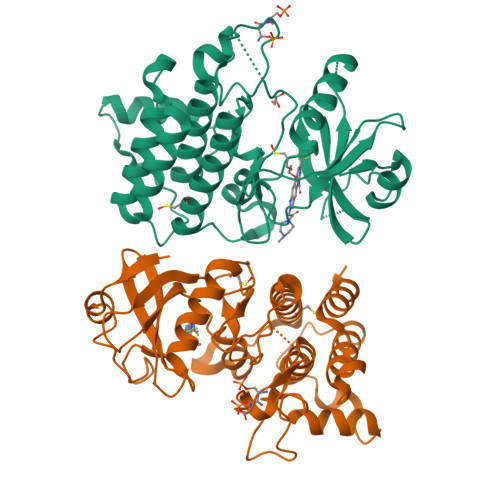
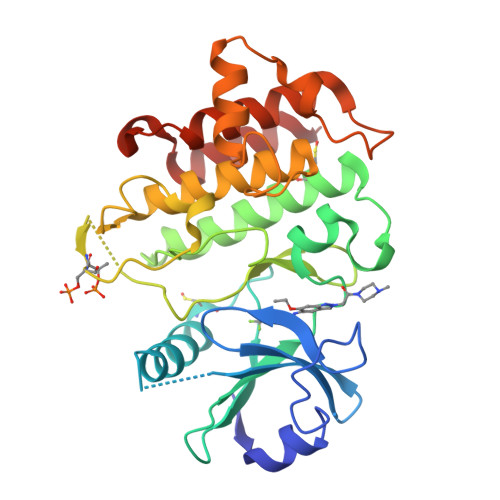
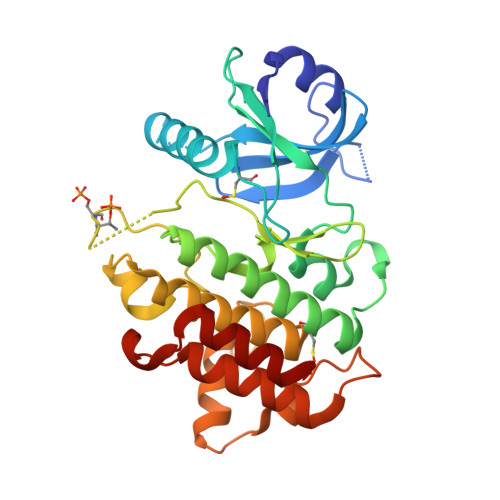
8ATB, 8ATL, 8ATN, 8BR5, 8BR6, 8BR7 - PubMed Abstract:
Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a critical role in innate inflammatory processes. Here, we describe the discovery of two clinical candidate IRAK4 inhibitors, BAY1834845 (zabedosertib) and BAY1830839 , starting from a high-throughput screening hit derived from Bayer's compound library. By exploiting binding site features distinct to IRAK4 using an in-house docking model, liabilities of the original hit could surprisingly be overcome to confer both candidates with a unique combination of good potency and selectivity. Favorable DMPK profiles and activity in animal inflammation models led to the selection of these two compounds for clinical development in patients.
Organizational Affiliation:
Bayer AG, Research & Development, Pharmaceuticals, 13353 Berlin, Germany.