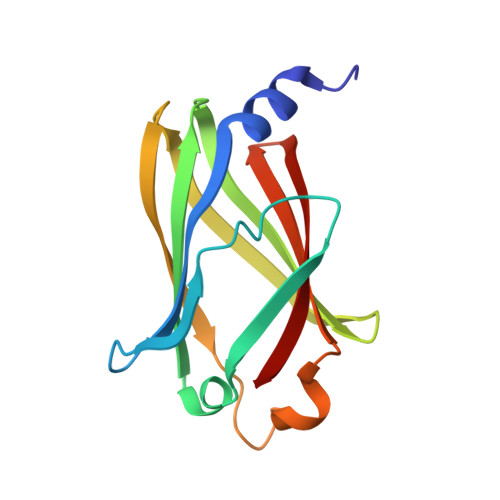
Stabilization of the RAS:PDE6D Complex Is a Novel Strategy to Inhibit RAS Signaling.
Yelland, T., Garcia, E., Parry, C., Kowalczyk, D., Wojnowska, M., Gohlke, A., Zalar, M., Cameron, K., Goodwin, G., Yu, Q., Zhu, P.C., ElMaghloob, Y., Pugliese, A., Archibald, L., Jamieson, A., Chen, Y.X., McArthur, D., Bower, J., Ismail, S.(2022) J Med Chem 65: 1898-1914
- PubMed: 35104933
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01265
- Primary Citation of Related Structures:
7Q9Q, 7Q9R, 7Q9S, 7Q9U, 7QF9, 7QJK - PubMed Abstract:
RAS is a major anticancer drug target which requires membrane localization to activate downstream signal transduction. The direct inhibition of RAS has proven to be challenging. Here, we present a novel strategy for targeting RAS by stabilizing its interaction with the prenyl-binding protein PDE6D and disrupting its localization. Using rationally designed RAS point mutations, we were able to stabilize the RAS:PDE6D complex by increasing the affinity of RAS for PDE6D, which resulted in the redirection of RAS to the cytoplasm and the primary cilium and inhibition of oncogenic RAS/ERK signaling. We developed an SPR fragment screening and identified fragments that bind at the KRAS:PDE6D interface, as shown through cocrystal structures. Finally, we show that the stoichiometric ratios of KRAS:PDE6D vary in different cell lines, suggesting that the impact of this strategy might be cell-type-dependent. This study forms the foundation from which a potential anticancer small-molecule RAS:PDE6D complex stabilizer could be developed.
- CRUK Beatson Institute, Glasgow G61 1BD, United Kingdom.
Organizational Affiliation: