Analogs of TIQ-A as inhibitors of human mono-ADP-ribosylating PARPs.
Maksimainen, M.M., Murthy, S., Sowa, S.T., Galera-Prat, A., Rolina, E., Heiskanen, J.P., Lehtio, L.(2021) Bioorg Med Chem 52: 116511-116511
- PubMed: 34801828
- DOI: https://doi.org/10.1016/j.bmc.2021.116511
- Primary Citation of Related Structures:
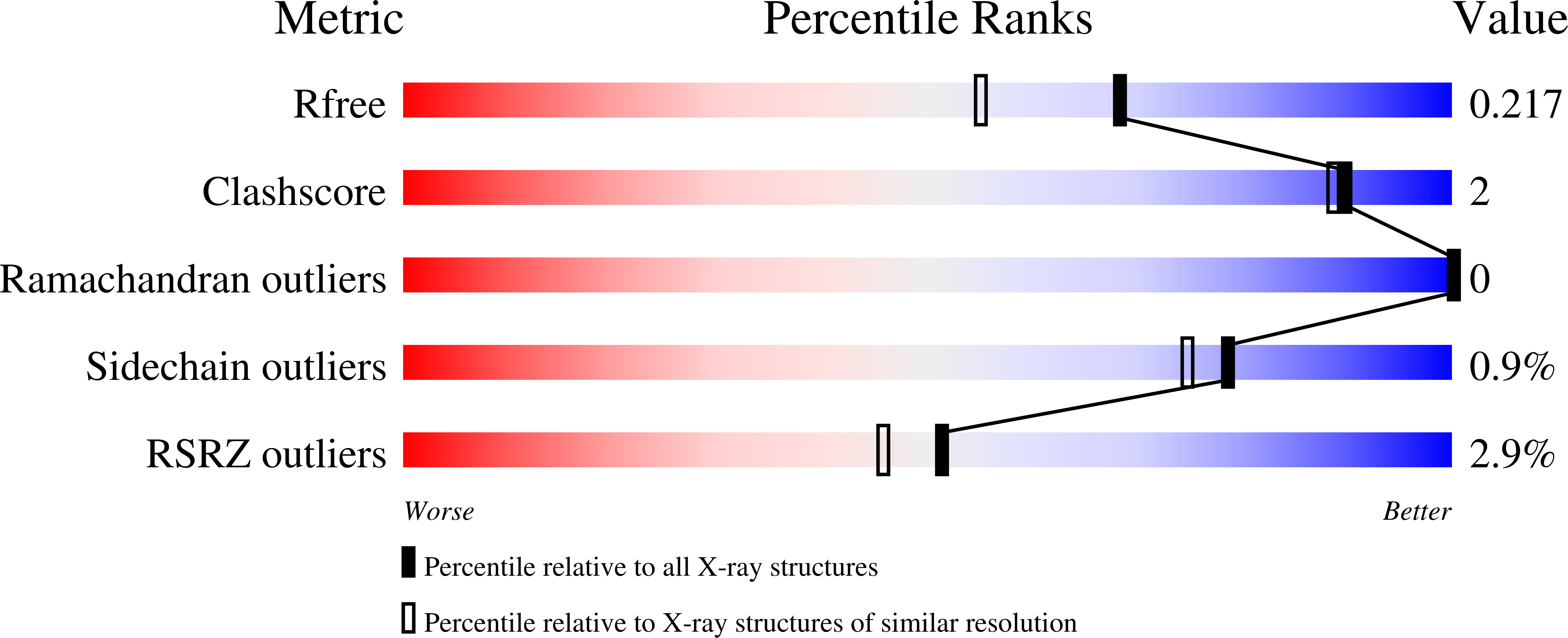
7OLJ, 7OM1, 7OMC, 7OQQ, 7OSP, 7OSS, 7OSX, 7OTF, 7OTH, 7OUW, 7OUX - PubMed Abstract:
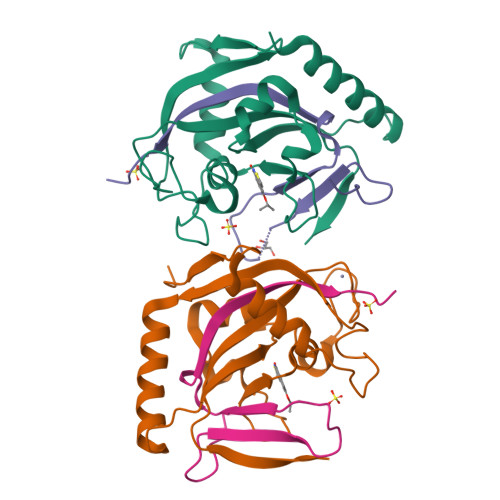
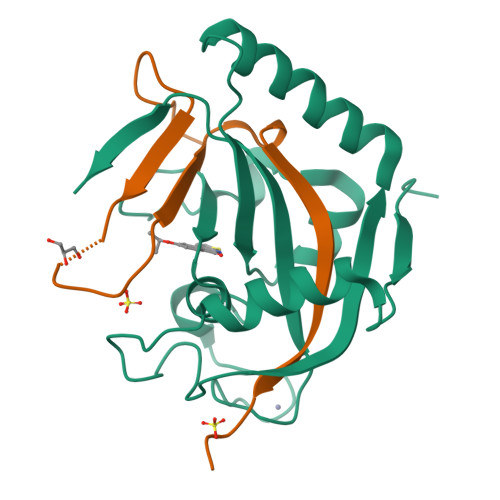
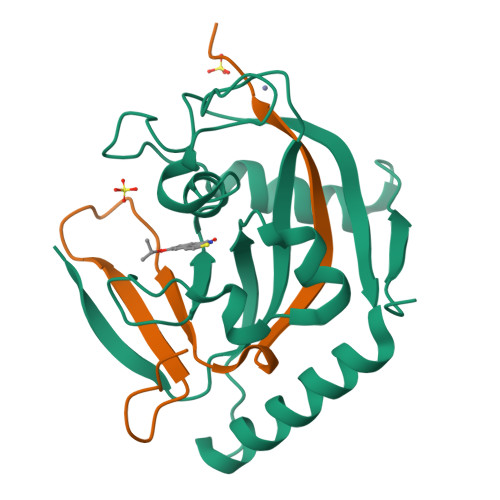
The scaffold of TIQ-A, a previously known inhibitor of human poly-ADP-ribosyltransferase PARP1, was utilized to develop inhibitors against human mono-ADP-ribosyltransferases through structure-guided design and activity profiling. By supplementing the TIQ-A scaffold with small structural changes, based on a PARP10 inhibitor OUL35, selectivity changed from poly-ADP-ribosyltransferases towards mono-ADP-ribosyltransferases. Binding modes of analogs were experimentally verified by determining complex crystal structures with mono-ADP-ribosyltransferase PARP15 and with poly-ADP-ribosyltransferase TNKS2. The best analogs of the study achieved 10-20-fold selectivity towards mono-ADP-ribosyltransferases PARP10 and PARP15 while maintaining micromolar potencies. The work demonstrates a route to differentiate compound selectivity between mono- and poly-ribosyltransferases of the human ARTD family.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Oulu, Finland.