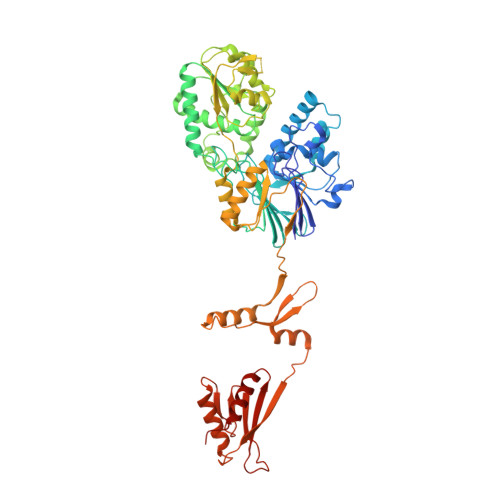
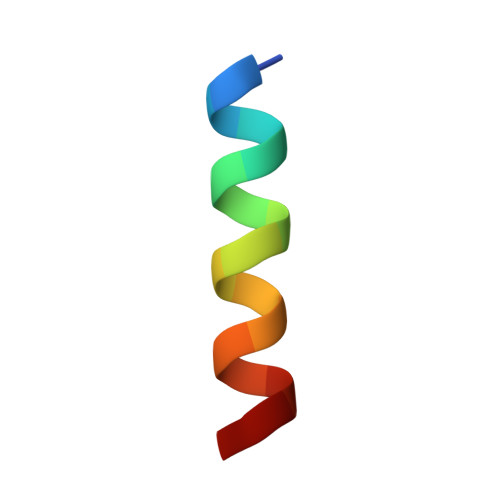
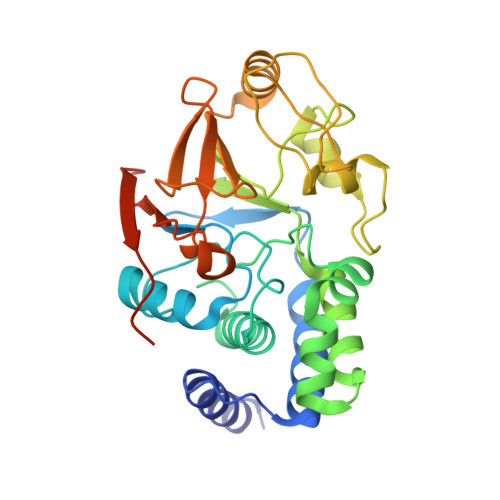
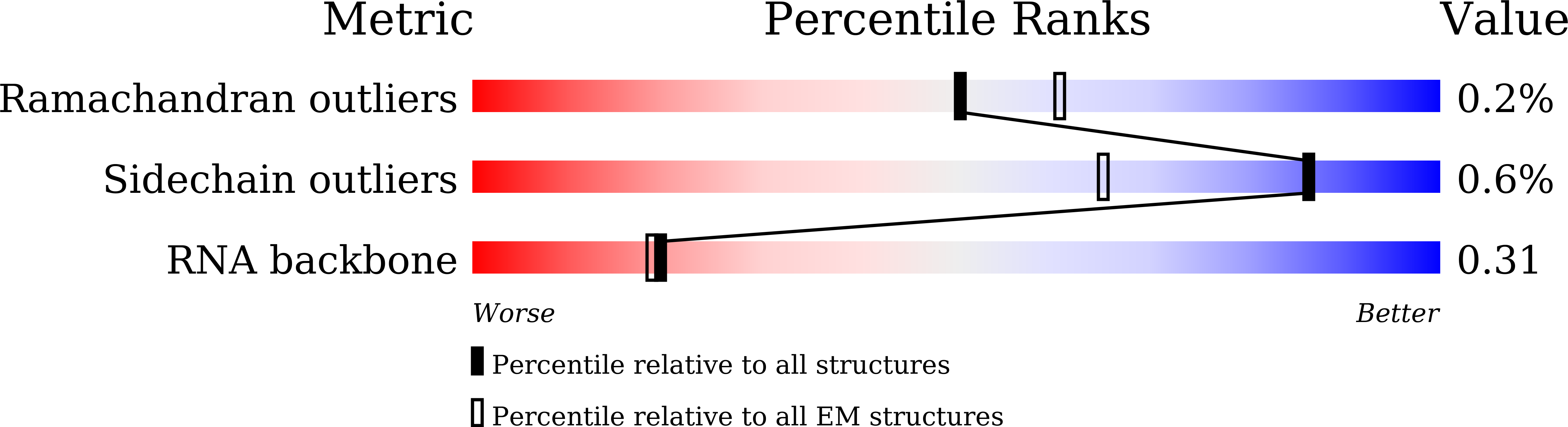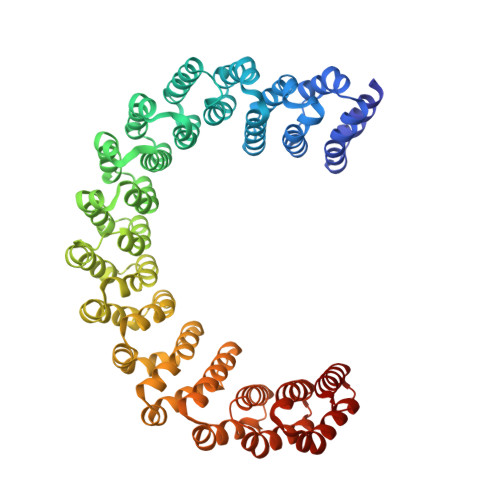Proteins 31 Nucleic Acids / Hybrid 3
Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by: Sequence | 3D Structure
Entity ID: 10 Molecule Chains Sequence Length Organism Details Image Unknown2 J [auth M] 17 Homo sapiens Mutation(s) : 0 Sequence Annotations
Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Entity ID: 13 Molecule Chains Sequence Length Organism Details Image Unknown M [auth U] 27 Homo sapiens Mutation(s) : 0 Sequence Annotations
Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure Find similar proteins by:
Sequence
(by identity cutoff) |
3D Structure