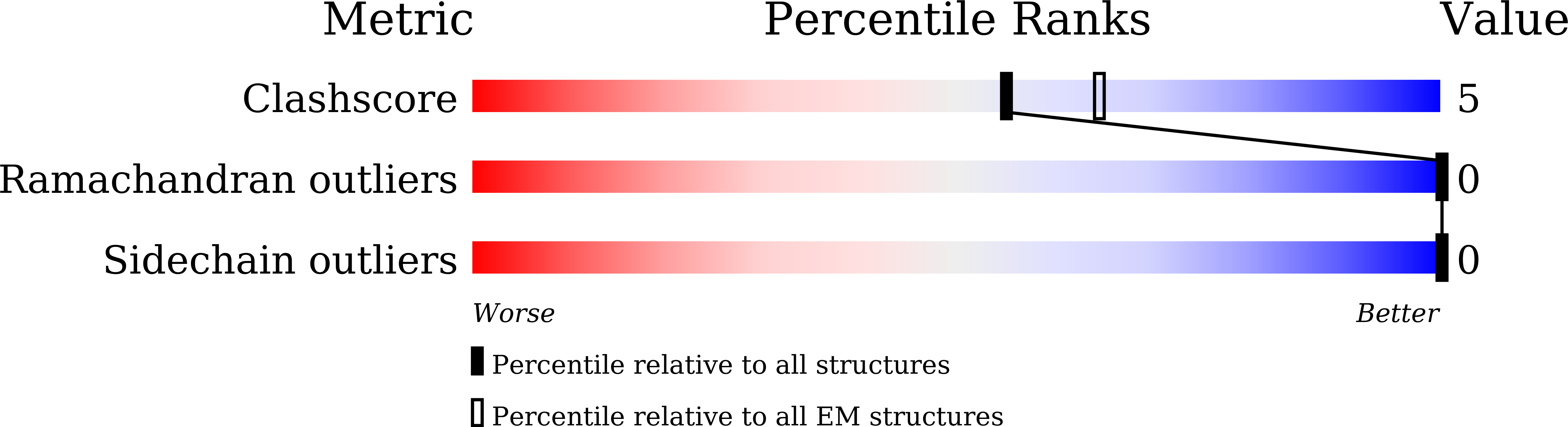GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Huang, S., Xu, P., Shen, D.D., Simon, I.A., Mao, C., Tan, Y., Zhang, H., Harpsoe, K., Li, H., Zhang, Y., You, C., Yu, X., Jiang, Y., Zhang, Y., Gloriam, D.E., Xu, H.E.(2022) Mol Cell 82: 2681-2695.e6
- PubMed: 35714614
- DOI: https://doi.org/10.1016/j.molcel.2022.05.031
- Primary Citation of Related Structures:
7XT8, 7XT9, 7XTA, 7XTB, 7XTC - PubMed Abstract:
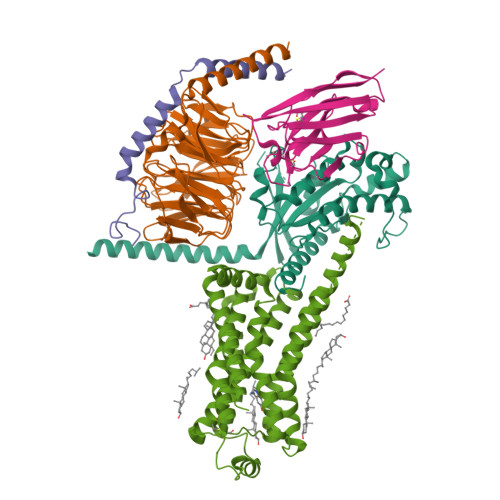
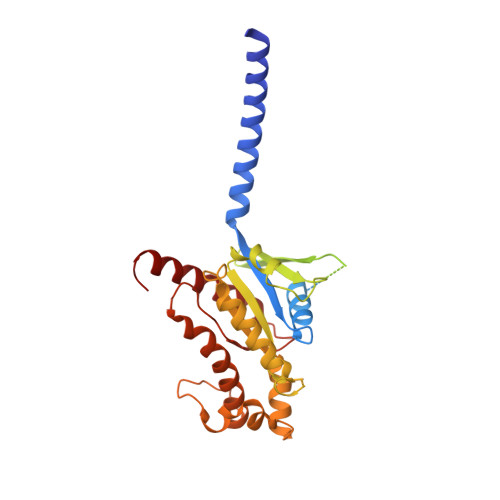
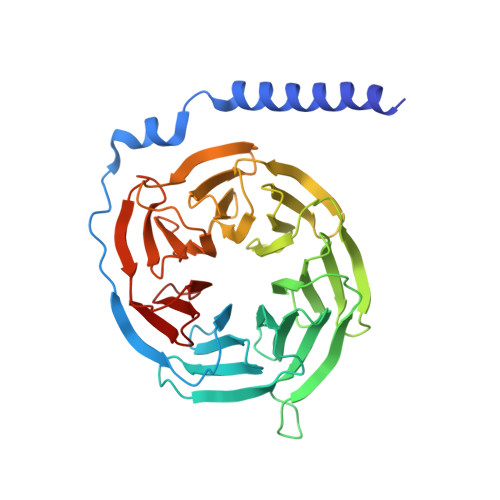
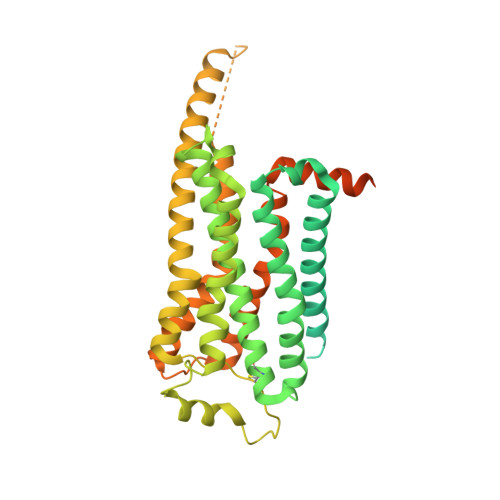
Serotonin (or 5-hydroxytryptamine, 5-HT) is an important neurotransmitter that activates 12 different G protein-coupled receptors (GPCRs) through selective coupling of G s , G i, or G q proteins. The structural basis for G protein subtype selectivity by these GPCRs remains elusive. Here, we report the structures of the serotonin receptors 5-HT 4 , 5-HT 6 , and 5-HT 7 with G s , and 5-HT 4 with G i1 . The structures reveal that transmembrane helices TM5 and TM6 alternate lengths as a macro-switch to determine receptor's selectivity for G s and G i , respectively. We find that the macro-switch by the TM5-TM6 length is shared by class A GPCR-G protein structures. Furthermore, we discover specific residues within TM5 and TM6 that function as micro-switches to form specific interactions with G s or G i . Together, these results present a common mechanism of G s versus G i protein coupling selectivity or promiscuity by class A GPCRs and extend the basis of ligand recognition at serotonin receptors.
Organizational Affiliation:
The CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; University of Chinese Academy of Sciences, Beijing 100049, China; School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.